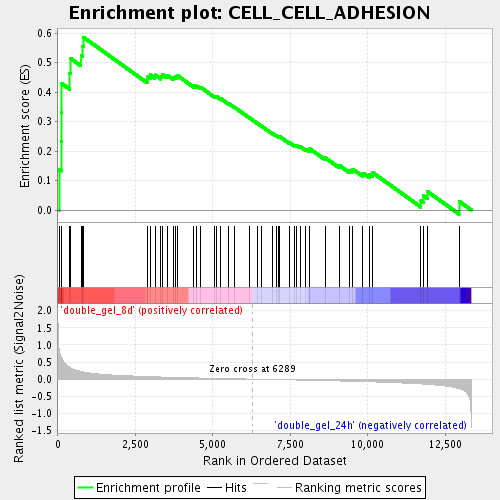
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | double_gel_and_single_gel_rma_expression_values_collapsed_to_symbols.class.cls #double_gel_8d_versus_double_gel_24h.class.cls #double_gel_8d_versus_double_gel_24h_repos |
| Phenotype | class.cls#double_gel_8d_versus_double_gel_24h_repos |
| Upregulated in class | double_gel_8d |
| GeneSet | CELL_CELL_ADHESION |
| Enrichment Score (ES) | 0.58592415 |
| Normalized Enrichment Score (NES) | 1.8257945 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.022948235 |
| FWER p-Value | 0.873 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | CLDN7 | CLDN7 Entrez, Source | claudin 7 | 36 | 0.888 | 0.1376 | Yes |
| 2 | MGP | MGP Entrez, Source | matrix Gla protein | 108 | 0.644 | 0.2340 | Yes |
| 3 | CLDN1 | CLDN1 Entrez, Source | claudin 1 | 118 | 0.623 | 0.3318 | Yes |
| 4 | PVRL2 | PVRL2 Entrez, Source | poliovirus receptor-related 2 (herpesvirus entry mediator B) | 126 | 0.616 | 0.4285 | Yes |
| 5 | APOA4 | APOA4 Entrez, Source | apolipoprotein A-IV | 383 | 0.346 | 0.4640 | Yes |
| 6 | CDH13 | CDH13 Entrez, Source | cadherin 13, H-cadherin (heart) | 417 | 0.328 | 0.5134 | Yes |
| 7 | CLDN3 | CLDN3 Entrez, Source | claudin 3 | 745 | 0.228 | 0.5248 | Yes |
| 8 | CX3CL1 | CX3CL1 Entrez, Source | chemokine (C-X3-C motif) ligand 1 | 796 | 0.215 | 0.5550 | Yes |
| 9 | LMO4 | LMO4 Entrez, Source | LIM domain only 4 | 824 | 0.209 | 0.5859 | Yes |
| 10 | ROBO2 | ROBO2 Entrez, Source | roundabout, axon guidance receptor, homolog 2 (Drosophila) | 2881 | 0.075 | 0.4430 | No |
| 11 | NLGN1 | NLGN1 Entrez, Source | neuroligin 1 | 2905 | 0.074 | 0.4530 | No |
| 12 | COL11A1 | COL11A1 Entrez, Source | collagen, type XI, alpha 1 | 2981 | 0.072 | 0.4587 | No |
| 13 | NPTN | NPTN Entrez, Source | neuroplastin | 3133 | 0.068 | 0.4580 | No |
| 14 | CNTN4 | CNTN4 Entrez, Source | contactin 4 | 3319 | 0.062 | 0.4539 | No |
| 15 | ATP2C1 | ATP2C1 Entrez, Source | ATPase, Ca++ transporting, type 2C, member 1 | 3361 | 0.061 | 0.4605 | No |
| 16 | ACVRL1 | ACVRL1 Entrez, Source | activin A receptor type II-like 1 | 3520 | 0.057 | 0.4576 | No |
| 17 | BCL10 | BCL10 Entrez, Source | B-cell CLL/lymphoma 10 | 3730 | 0.051 | 0.4500 | No |
| 18 | CLDN19 | CLDN19 Entrez, Source | claudin 19 | 3802 | 0.050 | 0.4526 | No |
| 19 | CD93 | CD93 Entrez, Source | CD93 molecule | 3868 | 0.048 | 0.4553 | No |
| 20 | CLDN8 | CLDN8 Entrez, Source | claudin 8 | 4382 | 0.036 | 0.4224 | No |
| 21 | THY1 | THY1 Entrez, Source | Thy-1 cell surface antigen | 4478 | 0.034 | 0.4207 | No |
| 22 | CDKN2A | CDKN2A Entrez, Source | cyclin-dependent kinase inhibitor 2A (melanoma, p16, inhibits CDK4) | 4610 | 0.032 | 0.4159 | No |
| 23 | PVRL1 | PVRL1 Entrez, Source | poliovirus receptor-related 1 (herpesvirus entry mediator C; nectin) | 5060 | 0.023 | 0.3857 | No |
| 24 | PKD1 | PKD1 Entrez, Source | polycystic kidney disease 1 (autosomal dominant) | 5117 | 0.022 | 0.3850 | No |
| 25 | CLDN14 | CLDN14 Entrez, Source | claudin 14 | 5251 | 0.020 | 0.3781 | No |
| 26 | CD164 | CD164 Entrez, Source | CD164 molecule, sialomucin | 5509 | 0.014 | 0.3610 | No |
| 27 | DLG1 | DLG1 Entrez, Source | discs, large homolog 1 (Drosophila) | 5702 | 0.011 | 0.3483 | No |
| 28 | TNF | TNF Entrez, Source | tumor necrosis factor (TNF superfamily, member 2) | 6181 | 0.001 | 0.3126 | No |
| 29 | LGALS7 | LGALS7 Entrez, Source | lectin, galactoside-binding, soluble, 7 (galectin 7) | 6454 | -0.003 | 0.2925 | No |
| 30 | BMP1 | BMP1 Entrez, Source | bone morphogenetic protein 1 | 6563 | -0.005 | 0.2852 | No |
| 31 | ROBO1 | ROBO1 Entrez, Source | roundabout, axon guidance receptor, homolog 1 (Drosophila) | 6932 | -0.011 | 0.2592 | No |
| 32 | SYK | SYK Entrez, Source | spleen tyrosine kinase | 7042 | -0.013 | 0.2531 | No |
| 33 | AMIGO2 | AMIGO2 Entrez, Source | adhesion molecule with Ig-like domain 2 | 7133 | -0.014 | 0.2486 | No |
| 34 | CALCA | CALCA Entrez, Source | calcitonin/calcitonin-related polypeptide, alpha | 7138 | -0.014 | 0.2506 | No |
| 35 | CDK5R1 | CDK5R1 Entrez, Source | cyclin-dependent kinase 5, regulatory subunit 1 (p35) | 7459 | -0.021 | 0.2298 | No |
| 36 | AMIGO3 | AMIGO3 Entrez, Source | adhesion molecule with Ig-like domain 3 | 7637 | -0.024 | 0.2203 | No |
| 37 | CLDN11 | CLDN11 Entrez, Source | claudin 11 (oligodendrocyte transmembrane protein) | 7693 | -0.025 | 0.2201 | No |
| 38 | EMCN | EMCN Entrez, Source | endomucin | 7816 | -0.028 | 0.2153 | No |
| 39 | CLDN4 | CLDN4 Entrez, Source | claudin 4 | 8005 | -0.031 | 0.2061 | No |
| 40 | CLDN5 | CLDN5 Entrez, Source | claudin 5 (transmembrane protein deleted in velocardiofacial syndrome) | 8120 | -0.034 | 0.2029 | No |
| 41 | ITGB1 | ITGB1 Entrez, Source | integrin, beta 1 (fibronectin receptor, beta polypeptide, antigen CD29 includes MDF2, MSK12) | 8134 | -0.034 | 0.2073 | No |
| 42 | CD47 | CD47 Entrez, Source | CD47 molecule | 8623 | -0.044 | 0.1775 | No |
| 43 | REG3A | REG3A Entrez, Source | regenerating islet-derived 3 alpha | 9086 | -0.054 | 0.1512 | No |
| 44 | NINJ2 | NINJ2 Entrez, Source | ninjurin 2 | 9422 | -0.061 | 0.1357 | No |
| 45 | NF2 | NF2 Entrez, Source | neurofibromin 2 (bilateral acoustic neuroma) | 9513 | -0.063 | 0.1389 | No |
| 46 | NCAM2 | NCAM2 Entrez, Source | neural cell adhesion molecule 2 | 9845 | -0.072 | 0.1254 | No |
| 47 | CLDN16 | CLDN16 Entrez, Source | claudin 16 | 10062 | -0.078 | 0.1215 | No |
| 48 | ITGB2 | ITGB2 Entrez, Source | integrin, beta 2 (complement component 3 receptor 3 and 4 subunit) | 10160 | -0.081 | 0.1269 | No |
| 49 | GTPBP4 | GTPBP4 Entrez, Source | GTP binding protein 4 | 11715 | -0.139 | 0.0320 | No |
| 50 | RASA1 | RASA1 Entrez, Source | RAS p21 protein activator (GTPase activating protein) 1 | 11793 | -0.144 | 0.0489 | No |
| 51 | PTEN | PTEN Entrez, Source | phosphatase and tensin homolog (mutated in multiple advanced cancers 1) | 11925 | -0.152 | 0.0630 | No |
| 52 | EGFR | EGFR Entrez, Source | epidermal growth factor receptor (erythroblastic leukemia viral (v-erb-b) oncogene homolog, avian) | 12946 | -0.276 | 0.0298 | No |