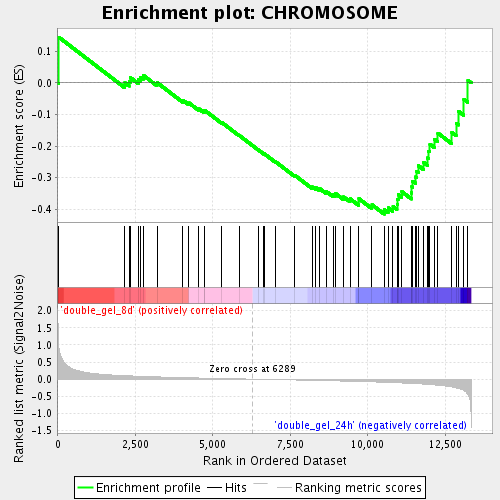
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | double_gel_and_single_gel_rma_expression_values_collapsed_to_symbols.class.cls #double_gel_8d_versus_double_gel_24h.class.cls #double_gel_8d_versus_double_gel_24h_repos |
| Phenotype | class.cls#double_gel_8d_versus_double_gel_24h_repos |
| Upregulated in class | double_gel_24h |
| GeneSet | CHROMOSOME |
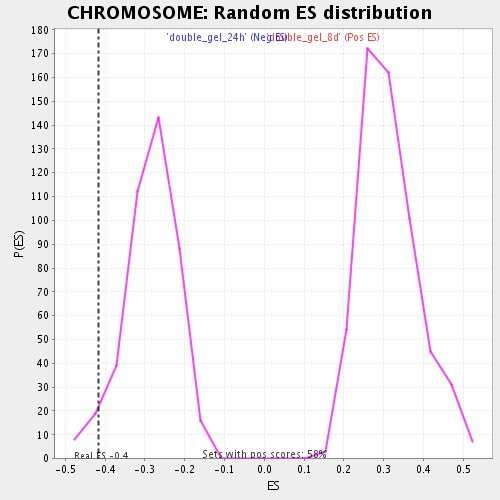
| Enrichment Score (ES) | -0.41528833 |
| Normalized Enrichment Score (NES) | -1.4521024 |
| Nominal p-value | 0.044705883 |
| FDR q-value | 0.25006285 |
| FWER p-Value | 1.0 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | MAF | MAF Entrez, Source | v-maf musculoaponeurotic fibrosarcoma oncogene homolog (avian) | 31 | 0.935 | 0.1448 | No |
| 2 | RB1 | RB1 Entrez, Source | retinoblastoma 1 (including osteosarcoma) | 2162 | 0.101 | 0.0004 | No |
| 3 | BUB1B | BUB1B Entrez, Source | BUB1 budding uninhibited by benzimidazoles 1 homolog beta (yeast) | 2298 | 0.096 | 0.0052 | No |
| 4 | HMGB1 | HMGB1 Entrez, Source | high-mobility group box 1 | 2337 | 0.094 | 0.0171 | No |
| 5 | XRCC4 | XRCC4 Entrez, Source | X-ray repair complementing defective repair in Chinese hamster cells 4 | 2599 | 0.083 | 0.0106 | No |
| 6 | TERF2IP | TERF2IP Entrez, Source | telomeric repeat binding factor 2, interacting protein | 2663 | 0.081 | 0.0187 | No |
| 7 | JUNB | JUNB Entrez, Source | jun B proto-oncogene | 2752 | 0.078 | 0.0244 | No |
| 8 | TUBG1 | TUBG1 Entrez, Source | tubulin, gamma 1 | 3200 | 0.066 | 0.0011 | No |
| 9 | REPIN1 | REPIN1 Entrez, Source | replication initiator 1 | 4032 | 0.044 | -0.0545 | No |
| 10 | TMPO | TMPO Entrez, Source | thymopoietin | 4229 | 0.039 | -0.0632 | No |
| 11 | ATRX | ATRX Entrez, Source | alpha thalassemia/mental retardation syndrome X-linked (RAD54 homolog, S. cerevisiae) | 4548 | 0.033 | -0.0819 | No |
| 12 | SC65 | SC65 Entrez, Source | - | 4719 | 0.030 | -0.0900 | No |
| 13 | PSEN2 | PSEN2 Entrez, Source | presenilin 2 (Alzheimer disease 4) | 4726 | 0.030 | -0.0858 | No |
| 14 | JUND | JUND Entrez, Source | jun D proto-oncogene | 5289 | 0.019 | -0.1251 | No |
| 15 | BIRC5 | BIRC5 Entrez, Source | baculoviral IAP repeat-containing 5 (survivin) | 5848 | 0.008 | -0.1658 | No |
| 16 | NPM2 | NPM2 Entrez, Source | nucleophosmin/nucleoplasmin, 2 | 6472 | -0.003 | -0.2122 | No |
| 17 | JUN | JUN Entrez, Source | jun oncogene | 6649 | -0.006 | -0.2245 | No |
| 18 | RGS12 | RGS12 Entrez, Source | regulator of G-protein signalling 12 | 6660 | -0.007 | -0.2242 | No |
| 19 | TOP1 | TOP1 Entrez, Source | topoisomerase (DNA) I | 7017 | -0.013 | -0.2490 | No |
| 20 | APC | APC Entrez, Source | adenomatosis polyposis coli | 7641 | -0.024 | -0.2921 | No |
| 21 | TTN | TTN Entrez, Source | titin | 8210 | -0.035 | -0.3292 | No |
| 22 | TNP1 | TNP1 Entrez, Source | transition protein 1 (during histone to protamine replacement) | 8326 | -0.038 | -0.3319 | No |
| 23 | UBE2I | UBE2I Entrez, Source | ubiquitin-conjugating enzyme E2I (UBC9 homolog, yeast) | 8446 | -0.040 | -0.3346 | No |
| 24 | STAG3 | STAG3 Entrez, Source | stromal antigen 3 | 8660 | -0.045 | -0.3436 | No |
| 25 | H2AFY | H2AFY Entrez, Source | H2A histone family, member Y | 8887 | -0.050 | -0.3528 | No |
| 26 | DCTN2 | DCTN2 Entrez, Source | dynactin 2 (p50) | 8962 | -0.051 | -0.3503 | No |
| 27 | BUB3 | BUB3 Entrez, Source | BUB3 budding uninhibited by benzimidazoles 3 homolog (yeast) | 9220 | -0.056 | -0.3608 | No |
| 28 | ZW10 | ZW10 Entrez, Source | ZW10, kinetochore associated, homolog (Drosophila) | 9428 | -0.061 | -0.3668 | No |
| 29 | RFC1 | RFC1 Entrez, Source | replication factor C (activator 1) 1, 145kDa | 9713 | -0.068 | -0.3774 | No |
| 30 | KLHDC3 | KLHDC3 Entrez, Source | kelch domain containing 3 | 9714 | -0.068 | -0.3666 | No |
| 31 | HMGN1 | HMGN1 Entrez, Source | high-mobility group nucleosome binding domain 1 | 10136 | -0.080 | -0.3857 | No |
| 32 | CHEK1 | CHEK1 Entrez, Source | CHK1 checkpoint homolog (S. pombe) | 10530 | -0.091 | -0.4010 | Yes |
| 33 | SMC1A | SMC1A Entrez, Source | structural maintenance of chromosomes 1A | 10658 | -0.096 | -0.3955 | Yes |
| 34 | SUGT1 | SUGT1 Entrez, Source | SGT1, suppressor of G2 allele of SKP1 (S. cerevisiae) | 10813 | -0.101 | -0.3911 | Yes |
| 35 | PAM | PAM Entrez, Source | peptidylglycine alpha-amidating monooxygenase | 10944 | -0.106 | -0.3842 | Yes |
| 36 | SMC4 | SMC4 Entrez, Source | structural maintenance of chromosomes 4 | 10956 | -0.107 | -0.3682 | Yes |
| 37 | CLASP1 | CLASP1 Entrez, Source | cytoplasmic linker associated protein 1 | 10986 | -0.108 | -0.3535 | Yes |
| 38 | LRPPRC | LRPPRC Entrez, Source | leucine-rich PPR-motif containing | 11099 | -0.112 | -0.3443 | Yes |
| 39 | TOP2A | TOP2A Entrez, Source | topoisomerase (DNA) II alpha 170kDa | 11410 | -0.125 | -0.3479 | Yes |
| 40 | RAN | RAN Entrez, Source | RAN, member RAS oncogene family | 11423 | -0.126 | -0.3290 | Yes |
| 41 | PCNA | PCNA Entrez, Source | proliferating cell nuclear antigen | 11449 | -0.127 | -0.3109 | Yes |
| 42 | TINF2 | TINF2 Entrez, Source | TERF1 (TRF1)-interacting nuclear factor 2 | 11547 | -0.131 | -0.2976 | Yes |
| 43 | PSEN1 | PSEN1 Entrez, Source | presenilin 1 (Alzheimer disease 3) | 11576 | -0.132 | -0.2789 | Yes |
| 44 | ZWINT | ZWINT Entrez, Source | ZW10 interactor | 11637 | -0.135 | -0.2621 | Yes |
| 45 | PAFAH1B1 | PAFAH1B1 Entrez, Source | platelet-activating factor acetylhydrolase, isoform Ib, alpha subunit 45kDa | 11805 | -0.145 | -0.2519 | Yes |
| 46 | RFC2 | RFC2 Entrez, Source | replication factor C (activator 1) 2, 40kDa | 11931 | -0.152 | -0.2374 | Yes |
| 47 | NUFIP1 | NUFIP1 Entrez, Source | nuclear fragile X mental retardation protein interacting protein 1 | 11965 | -0.155 | -0.2156 | Yes |
| 48 | RPA2 | RPA2 Entrez, Source | replication protein A2, 32kDa | 12006 | -0.158 | -0.1937 | Yes |
| 49 | MKI67IP | MKI67IP Entrez, Source | MKI67 (FHA domain) interacting nucleolar phosphoprotein | 12147 | -0.168 | -0.1779 | Yes |
| 50 | DNMT3A | DNMT3A Entrez, Source | DNA (cytosine-5-)-methyltransferase 3 alpha | 12263 | -0.177 | -0.1587 | Yes |
| 51 | RFC3 | RFC3 Entrez, Source | replication factor C (activator 1) 3, 38kDa | 12712 | -0.226 | -0.1569 | Yes |
| 52 | TIMELESS | TIMELESS Entrez, Source | timeless homolog (Drosophila) | 12855 | -0.255 | -0.1275 | Yes |
| 53 | KIF22 | KIF22 Entrez, Source | kinesin family member 22 | 12933 | -0.273 | -0.0903 | Yes |
| 54 | MCM7 | MCM7 Entrez, Source | MCM7 minichromosome maintenance deficient 7 (S. cerevisiae) | 13101 | -0.328 | -0.0514 | Yes |
| 55 | APTX | APTX Entrez, Source | aprataxin | 13232 | -0.441 | 0.0083 | Yes |