Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | double_gel_and_single_gel_rma_expression_values_collapsed_to_symbols.class.cls #double_gel_8d_versus_double_gel_24h.class.cls #double_gel_8d_versus_double_gel_24h_repos |
| Phenotype | class.cls#double_gel_8d_versus_double_gel_24h_repos |
| Upregulated in class | double_gel_24h |
| GeneSet | CMV_HCMV_TIMECOURSE_18HRS_UP |
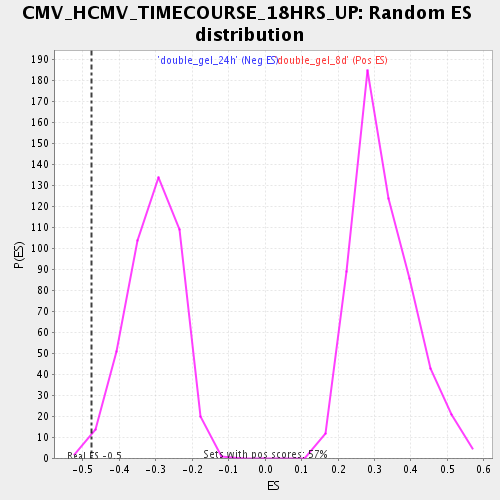
| Enrichment Score (ES) | -0.47785518 |
| Normalized Enrichment Score (NES) | -1.5601147 |
| Nominal p-value | 0.013793103 |
| FDR q-value | 0.19654295 |
| FWER p-Value | 1.0 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | TSC22D3 | TSC22D3 Entrez, Source | TSC22 domain family, member 3 | 257 | 0.432 | 0.0570 | No |
| 2 | NEU1 | NEU1 Entrez, Source | sialidase 1 (lysosomal sialidase) | 866 | 0.201 | 0.0468 | No |
| 3 | PTGER2 | PTGER2 Entrez, Source | prostaglandin E receptor 2 (subtype EP2), 53kDa | 1419 | 0.141 | 0.0302 | No |
| 4 | OLFM1 | OLFM1 Entrez, Source | olfactomedin 1 | 1771 | 0.118 | 0.0247 | No |
| 5 | BRAF | BRAF Entrez, Source | v-raf murine sarcoma viral oncogene homolog B1 | 2605 | 0.083 | -0.0232 | No |
| 6 | PDLIM3 | PDLIM3 Entrez, Source | PDZ and LIM domain 3 | 3680 | 0.053 | -0.0946 | No |
| 7 | DYNC1I1 | DYNC1I1 Entrez, Source | dynein, cytoplasmic 1, intermediate chain 1 | 4021 | 0.044 | -0.1124 | No |
| 8 | DMN | DMN Entrez, Source | desmuslin | 4837 | 0.027 | -0.1689 | No |
| 9 | CCNC | CCNC Entrez, Source | cyclin C | 4913 | 0.026 | -0.1700 | No |
| 10 | ISL1 | ISL1 Entrez, Source | ISL1 transcription factor, LIM/homeodomain, (islet-1) | 5512 | 0.014 | -0.2124 | No |
| 11 | WDR47 | WDR47 Entrez, Source | WD repeat domain 47 | 5564 | 0.013 | -0.2139 | No |
| 12 | DLG1 | DLG1 Entrez, Source | discs, large homolog 1 (Drosophila) | 5702 | 0.011 | -0.2223 | No |
| 13 | PDE4D | PDE4D Entrez, Source | phosphodiesterase 4D, cAMP-specific (phosphodiesterase E3 dunce homolog, Drosophila) | 5890 | 0.007 | -0.2350 | No |
| 14 | RCHY1 | RCHY1 Entrez, Source | ring finger and CHY zinc finger domain containing 1 | 5935 | 0.006 | -0.2372 | No |
| 15 | DR1 | DR1 Entrez, Source | down-regulator of transcription 1, TBP-binding (negative cofactor 2) | 6831 | -0.009 | -0.3029 | No |
| 16 | SSR1 | SSR1 Entrez, Source | signal sequence receptor, alpha (translocon-associated protein alpha) | 8002 | -0.031 | -0.3853 | No |
| 17 | KCNS3 | KCNS3 Entrez, Source | potassium voltage-gated channel, delayed-rectifier, subfamily S, member 3 | 8138 | -0.034 | -0.3895 | No |
| 18 | CRLF3 | CRLF3 Entrez, Source | cytokine receptor-like factor 3 | 8904 | -0.050 | -0.4382 | No |
| 19 | SON | SON Entrez, Source | SON DNA binding protein | 9229 | -0.057 | -0.4525 | No |
| 20 | TESK2 | TESK2 Entrez, Source | testis-specific kinase 2 | 9339 | -0.059 | -0.4503 | No |
| 21 | PPM1A | PPM1A Entrez, Source | protein phosphatase 1A (formerly 2C), magnesium-dependent, alpha isoform | 9447 | -0.062 | -0.4474 | No |
| 22 | STCH | STCH Entrez, Source | stress 70 protein chaperone, microsome-associated, 60kDa | 9623 | -0.066 | -0.4489 | No |
| 23 | POLE3 | POLE3 Entrez, Source | polymerase (DNA directed), epsilon 3 (p17 subunit) | 9639 | -0.066 | -0.4384 | No |
| 24 | RNF2 | RNF2 Entrez, Source | ring finger protein 2 | 9684 | -0.067 | -0.4297 | No |
| 25 | RFK | RFK Entrez, Source | riboflavin kinase | 10120 | -0.080 | -0.4484 | No |
| 26 | MEF2D | MEF2D Entrez, Source | MADS box transcription enhancer factor 2, polypeptide D (myocyte enhancer factor 2D) | 10414 | -0.088 | -0.4549 | No |
| 27 | EP300 | EP300 Entrez, Source | E1A binding protein p300 | 10720 | -0.098 | -0.4606 | Yes |
| 28 | GATM | GATM Entrez, Source | glycine amidinotransferase (L-arginine:glycine amidinotransferase) | 10782 | -0.100 | -0.4475 | Yes |
| 29 | BRMS1 | BRMS1 Entrez, Source | breast cancer metastasis suppressor 1 | 11108 | -0.112 | -0.4521 | Yes |
| 30 | ANP32E | ANP32E Entrez, Source | acidic (leucine-rich) nuclear phosphoprotein 32 family, member E | 11114 | -0.113 | -0.4325 | Yes |
| 31 | TMF1 | TMF1 Entrez, Source | TATA element modulatory factor 1 | 11317 | -0.121 | -0.4264 | Yes |
| 32 | ARHGEF5 | ARHGEF5 Entrez, Source | Rho guanine nucleotide exchange factor (GEF) 5 | 11536 | -0.131 | -0.4197 | Yes |
| 33 | ATP6V1C1 | ATP6V1C1 Entrez, Source | ATPase, H+ transporting, lysosomal 42kDa, V1 subunit C1 | 11674 | -0.137 | -0.4058 | Yes |
| 34 | NUP153 | NUP153 Entrez, Source | nucleoporin 153kDa | 11979 | -0.156 | -0.4011 | Yes |
| 35 | RPS6KB1 | RPS6KB1 Entrez, Source | ribosomal protein S6 kinase, 70kDa, polypeptide 1 | 12220 | -0.173 | -0.3886 | Yes |
| 36 | TRIM23 | TRIM23 Entrez, Source | tripartite motif-containing 23 | 12278 | -0.177 | -0.3615 | Yes |
| 37 | ZBTB1 | ZBTB1 Entrez, Source | zinc finger and BTB domain containing 1 | 12279 | -0.177 | -0.3302 | Yes |
| 38 | GCH1 | GCH1 Entrez, Source | GTP cyclohydrolase 1 (dopa-responsive dystonia) | 12298 | -0.179 | -0.2998 | Yes |
| 39 | STAM2 | STAM2 Entrez, Source | signal transducing adaptor molecule (SH3 domain and ITAM motif) 2 | 12300 | -0.180 | -0.2682 | Yes |
| 40 | SLC12A2 | SLC12A2 Entrez, Source | solute carrier family 12 (sodium/potassium/chloride transporters), member 2 | 12512 | -0.201 | -0.2486 | Yes |
| 41 | CRY1 | CRY1 Entrez, Source | cryptochrome 1 (photolyase-like) | 12664 | -0.219 | -0.2212 | Yes |
| 42 | PPAT | PPAT Entrez, Source | phosphoribosyl pyrophosphate amidotransferase | 12680 | -0.221 | -0.1832 | Yes |
| 43 | CASP2 | CASP2 Entrez, Source | caspase 2, apoptosis-related cysteine peptidase (neural precursor cell expressed, developmentally down-regulated 2) | 13151 | -0.360 | -0.1549 | Yes |
| 44 | FNBP4 | FNBP4 Entrez, Source | formin binding protein 4 | 13248 | -0.477 | -0.0778 | Yes |
| 45 | GCLC | GCLC Entrez, Source | glutamate-cysteine ligase, catalytic subunit | 13249 | -0.480 | 0.0070 | Yes |