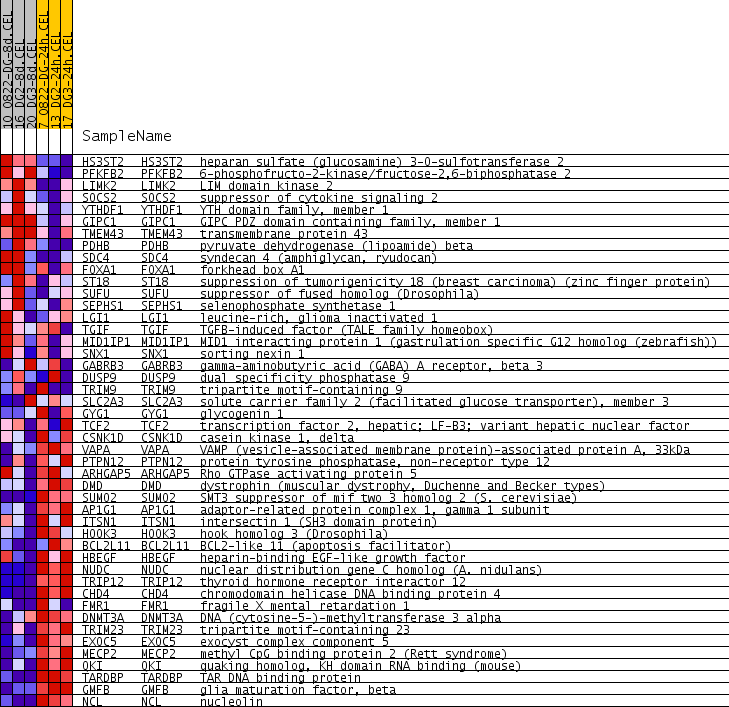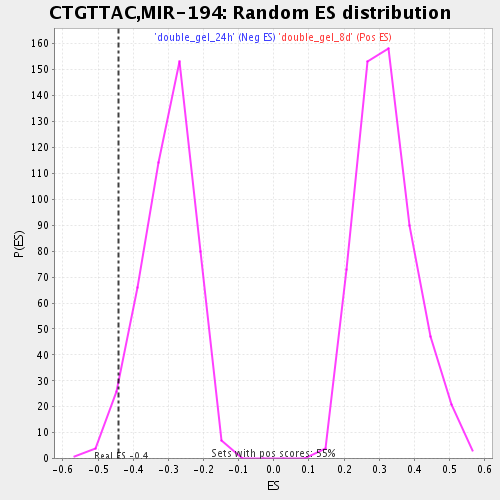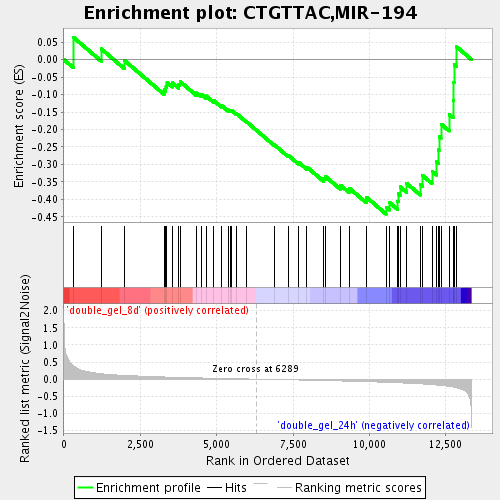
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | double_gel_and_single_gel_rma_expression_values_collapsed_to_symbols.class.cls #double_gel_8d_versus_double_gel_24h.class.cls #double_gel_8d_versus_double_gel_24h_repos |
| Phenotype | class.cls#double_gel_8d_versus_double_gel_24h_repos |
| Upregulated in class | double_gel_24h |
| GeneSet | CTGTTAC,MIR-194 |
| Enrichment Score (ES) | -0.44227955 |
| Normalized Enrichment Score (NES) | -1.4636905 |
| Nominal p-value | 0.042128604 |
| FDR q-value | 0.24810627 |
| FWER p-Value | 1.0 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | HS3ST2 | HS3ST2 Entrez, Source | heparan sulfate (glucosamine) 3-O-sulfotransferase 2 | 308 | 0.394 | 0.0641 | No |
| 2 | PFKFB2 | PFKFB2 Entrez, Source | 6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 2 | 1220 | 0.159 | 0.0309 | No |
| 3 | LIMK2 | LIMK2 Entrez, Source | LIM domain kinase 2 | 1982 | 0.109 | -0.0023 | No |
| 4 | SOCS2 | SOCS2 Entrez, Source | suppressor of cytokine signaling 2 | 3280 | 0.063 | -0.0858 | No |
| 5 | YTHDF1 | YTHDF1 Entrez, Source | YTH domain family, member 1 | 3340 | 0.062 | -0.0766 | No |
| 6 | GIPC1 | GIPC1 Entrez, Source | GIPC PDZ domain containing family, member 1 | 3372 | 0.061 | -0.0654 | No |
| 7 | TMEM43 | TMEM43 Entrez, Source | transmembrane protein 43 | 3546 | 0.056 | -0.0660 | No |
| 8 | PDHB | PDHB Entrez, Source | pyruvate dehydrogenase (lipoamide) beta | 3755 | 0.051 | -0.0704 | No |
| 9 | SDC4 | SDC4 Entrez, Source | syndecan 4 (amphiglycan, ryudocan) | 3799 | 0.050 | -0.0625 | No |
| 10 | FOXA1 | FOXA1 Entrez, Source | forkhead box A1 | 4345 | 0.037 | -0.0953 | No |
| 11 | ST18 | ST18 Entrez, Source | suppression of tumorigenicity 18 (breast carcinoma) (zinc finger protein) | 4497 | 0.034 | -0.0992 | No |
| 12 | SUFU | SUFU Entrez, Source | suppressor of fused homolog (Drosophila) | 4653 | 0.031 | -0.1040 | No |
| 13 | SEPHS1 | SEPHS1 Entrez, Source | selenophosphate synthetase 1 | 4894 | 0.026 | -0.1162 | No |
| 14 | LGI1 | LGI1 Entrez, Source | leucine-rich, glioma inactivated 1 | 5164 | 0.021 | -0.1318 | No |
| 15 | TGIF | TGIF Entrez, Source | TGFB-induced factor (TALE family homeobox) | 5378 | 0.017 | -0.1440 | No |
| 16 | MID1IP1 | MID1IP1 Entrez, Source | MID1 interacting protein 1 (gastrulation specific G12 homolog (zebrafish)) | 5440 | 0.016 | -0.1450 | No |
| 17 | SNX1 | SNX1 Entrez, Source | sorting nexin 1 | 5489 | 0.015 | -0.1453 | No |
| 18 | GABRB3 | GABRB3 Entrez, Source | gamma-aminobutyric acid (GABA) A receptor, beta 3 | 5640 | 0.012 | -0.1539 | No |
| 19 | DUSP9 | DUSP9 Entrez, Source | dual specificity phosphatase 9 | 5974 | 0.006 | -0.1777 | No |
| 20 | TRIM9 | TRIM9 Entrez, Source | tripartite motif-containing 9 | 6878 | -0.010 | -0.2434 | No |
| 21 | SLC2A3 | SLC2A3 Entrez, Source | solute carrier family 2 (facilitated glucose transporter), member 3 | 7341 | -0.019 | -0.2740 | No |
| 22 | GYG1 | GYG1 Entrez, Source | glycogenin 1 | 7679 | -0.025 | -0.2938 | No |
| 23 | TCF2 | TCF2 Entrez, Source | transcription factor 2, hepatic; LF-B3; variant hepatic nuclear factor | 7953 | -0.030 | -0.3077 | No |
| 24 | CSNK1D | CSNK1D Entrez, Source | casein kinase 1, delta | 8498 | -0.041 | -0.3394 | No |
| 25 | VAPA | VAPA Entrez, Source | VAMP (vesicle-associated membrane protein)-associated protein A, 33kDa | 8555 | -0.043 | -0.3342 | No |
| 26 | PTPN12 | PTPN12 Entrez, Source | protein tyrosine phosphatase, non-receptor type 12 | 9058 | -0.053 | -0.3602 | No |
| 27 | ARHGAP5 | ARHGAP5 Entrez, Source | Rho GTPase activating protein 5 | 9336 | -0.059 | -0.3679 | No |
| 28 | DMD | DMD Entrez, Source | dystrophin (muscular dystrophy, Duchenne and Becker types) | 9896 | -0.074 | -0.3936 | No |
| 29 | SUMO2 | SUMO2 Entrez, Source | SMT3 suppressor of mif two 3 homolog 2 (S. cerevisiae) | 10544 | -0.092 | -0.4220 | Yes |
| 30 | AP1G1 | AP1G1 Entrez, Source | adaptor-related protein complex 1, gamma 1 subunit | 10662 | -0.096 | -0.4095 | Yes |
| 31 | ITSN1 | ITSN1 Entrez, Source | intersectin 1 (SH3 domain protein) | 10914 | -0.105 | -0.4050 | Yes |
| 32 | HOOK3 | HOOK3 Entrez, Source | hook homolog 3 (Drosophila) | 10945 | -0.106 | -0.3837 | Yes |
| 33 | BCL2L11 | BCL2L11 Entrez, Source | BCL2-like 11 (apoptosis facilitator) | 11005 | -0.108 | -0.3641 | Yes |
| 34 | HBEGF | HBEGF Entrez, Source | heparin-binding EGF-like growth factor | 11219 | -0.117 | -0.3543 | Yes |
| 35 | NUDC | NUDC Entrez, Source | nuclear distribution gene C homolog (A. nidulans) | 11666 | -0.137 | -0.3575 | Yes |
| 36 | TRIP12 | TRIP12 Entrez, Source | thyroid hormone receptor interactor 12 | 11740 | -0.140 | -0.3319 | Yes |
| 37 | CHD4 | CHD4 Entrez, Source | chromodomain helicase DNA binding protein 4 | 12048 | -0.161 | -0.3194 | Yes |
| 38 | FMR1 | FMR1 Entrez, Source | fragile X mental retardation 1 | 12198 | -0.171 | -0.2927 | Yes |
| 39 | DNMT3A | DNMT3A Entrez, Source | DNA (cytosine-5-)-methyltransferase 3 alpha | 12263 | -0.177 | -0.2584 | Yes |
| 40 | TRIM23 | TRIM23 Entrez, Source | tripartite motif-containing 23 | 12278 | -0.177 | -0.2201 | Yes |
| 41 | EXOC5 | EXOC5 Entrez, Source | exocyst complex component 5 | 12342 | -0.183 | -0.1844 | Yes |
| 42 | MECP2 | MECP2 Entrez, Source | methyl CpG binding protein 2 (Rett syndrome) | 12609 | -0.212 | -0.1573 | Yes |
| 43 | QKI | QKI Entrez, Source | quaking homolog, KH domain RNA binding (mouse) | 12737 | -0.231 | -0.1158 | Yes |
| 44 | TARDBP | TARDBP Entrez, Source | TAR DNA binding protein | 12761 | -0.235 | -0.0655 | Yes |
| 45 | GMFB | GMFB Entrez, Source | glia maturation factor, beta | 12770 | -0.237 | -0.0137 | Yes |
| 46 | NCL | NCL Entrez, Source | nucleolin | 12857 | -0.256 | 0.0365 | Yes |