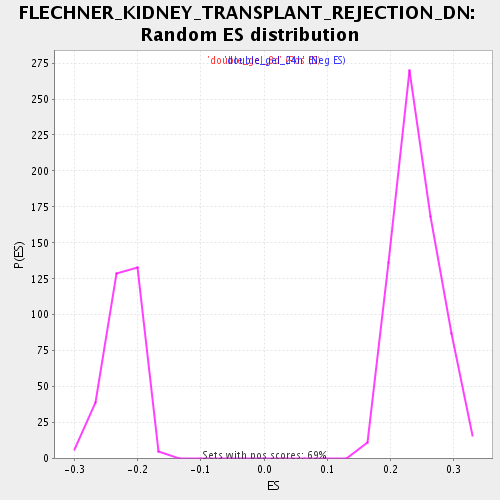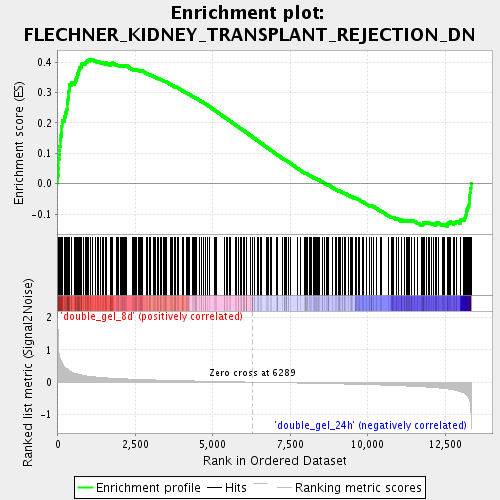
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | double_gel_and_single_gel_rma_expression_values_collapsed_to_symbols.class.cls #double_gel_8d_versus_double_gel_24h.class.cls #double_gel_8d_versus_double_gel_24h_repos |
| Phenotype | class.cls#double_gel_8d_versus_double_gel_24h_repos |
| Upregulated in class | double_gel_8d |
| GeneSet | FLECHNER_KIDNEY_TRANSPLANT_REJECTION_DN |
| Enrichment Score (ES) | 0.41022083 |
| Normalized Enrichment Score (NES) | 1.7028275 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.054057408 |
| FWER p-Value | 1.0 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | CYP17A1 | CYP17A1 Entrez, Source | cytochrome P450, family 17, subfamily A, polypeptide 1 | 2 | 1.623 | 0.0289 | Yes |
| 2 | SLC25A4 | SLC25A4 Entrez, Source | solute carrier family 25 (mitochondrial carrier; adenine nucleotide translocator), member 4 | 13 | 1.195 | 0.0495 | Yes |
| 3 | MAF | MAF Entrez, Source | v-maf musculoaponeurotic fibrosarcoma oncogene homolog (avian) | 31 | 0.935 | 0.0650 | Yes |
| 4 | DAB2 | DAB2 Entrez, Source | disabled homolog 2, mitogen-responsive phosphoprotein (Drosophila) | 32 | 0.925 | 0.0815 | Yes |
| 5 | CD9 | CD9 Entrez, Source | CD9 molecule | 41 | 0.871 | 0.0965 | Yes |
| 6 | ARMCX2 | ARMCX2 Entrez, Source | armadillo repeat containing, X-linked 2 | 51 | 0.809 | 0.1103 | Yes |
| 7 | CRYAB | CRYAB Entrez, Source | crystallin, alpha B | 62 | 0.756 | 0.1230 | Yes |
| 8 | GOT1 | GOT1 Entrez, Source | glutamic-oxaloacetic transaminase 1, soluble (aspartate aminotransferase 1) | 76 | 0.697 | 0.1345 | Yes |
| 9 | BBOX1 | BBOX1 Entrez, Source | butyrobetaine (gamma), 2-oxoglutarate dioxygenase (gamma-butyrobetaine hydroxylase) 1 | 78 | 0.695 | 0.1469 | Yes |
| 10 | LGALS2 | LGALS2 Entrez, Source | lectin, galactoside-binding, soluble, 2 (galectin 2) | 95 | 0.672 | 0.1577 | Yes |
| 11 | F2R | F2R Entrez, Source | coagulation factor II (thrombin) receptor | 114 | 0.625 | 0.1675 | Yes |
| 12 | PCK1 | PCK1 Entrez, Source | phosphoenolpyruvate carboxykinase 1 (soluble) | 127 | 0.610 | 0.1775 | Yes |
| 13 | AZGP1 | AZGP1 Entrez, Source | alpha-2-glycoprotein 1, zinc | 129 | 0.603 | 0.1882 | Yes |
| 14 | PAH | PAH Entrez, Source | phenylalanine hydroxylase | 133 | 0.597 | 0.1987 | Yes |
| 15 | BEXL1 | BEXL1 Entrez, Source | brain expressed X-linked-like 1 | 135 | 0.595 | 0.2093 | Yes |
| 16 | FBP1 | FBP1 Entrez, Source | fructose-1,6-bisphosphatase 1 | 203 | 0.484 | 0.2128 | Yes |
| 17 | USP2 | USP2 Entrez, Source | ubiquitin specific peptidase 2 | 207 | 0.480 | 0.2211 | Yes |
| 18 | FAH | FAH Entrez, Source | fumarylacetoacetate hydrolase (fumarylacetoacetase) | 232 | 0.457 | 0.2275 | Yes |
| 19 | GPX3 | GPX3 Entrez, Source | glutathione peroxidase 3 (plasma) | 242 | 0.449 | 0.2348 | Yes |
| 20 | DPYS | DPYS Entrez, Source | dihydropyrimidinase | 287 | 0.410 | 0.2388 | Yes |
| 21 | PRNP | PRNP Entrez, Source | prion protein (p27-30) (Creutzfeldt-Jakob disease, Gerstmann-Strausler-Scheinker syndrome, fatal familial insomnia) | 290 | 0.408 | 0.2459 | Yes |
| 22 | CDS1 | CDS1 Entrez, Source | CDP-diacylglycerol synthase (phosphatidate cytidylyltransferase) 1 | 299 | 0.402 | 0.2525 | Yes |
| 23 | ALDH6A1 | ALDH6A1 Entrez, Source | aldehyde dehydrogenase 6 family, member A1 | 300 | 0.402 | 0.2597 | Yes |
| 24 | DCI | DCI Entrez, Source | dodecenoyl-Coenzyme A delta isomerase (3,2 trans-enoyl-Coenzyme A isomerase) | 302 | 0.399 | 0.2668 | Yes |
| 25 | PC | PC Entrez, Source | pyruvate carboxylase | 322 | 0.380 | 0.2721 | Yes |
| 26 | RBP4 | RBP4 Entrez, Source | retinol binding protein 4, plasma | 323 | 0.378 | 0.2789 | Yes |
| 27 | SELENBP1 | SELENBP1 Entrez, Source | selenium binding protein 1 | 336 | 0.371 | 0.2846 | Yes |
| 28 | ECHS1 | ECHS1 Entrez, Source | enoyl Coenzyme A hydratase, short chain, 1, mitochondrial | 346 | 0.367 | 0.2905 | Yes |
| 29 | G6PC | G6PC Entrez, Source | glucose-6-phosphatase, catalytic subunit | 347 | 0.366 | 0.2970 | Yes |
| 30 | ECH1 | ECH1 Entrez, Source | enoyl Coenzyme A hydratase 1, peroxisomal | 350 | 0.365 | 0.3034 | Yes |
| 31 | CYB5A | CYB5A Entrez, Source | cytochrome b5 type A (microsomal) | 359 | 0.359 | 0.3092 | Yes |
| 32 | HSPA4 | HSPA4 Entrez, Source | heat shock 70kDa protein 4 | 361 | 0.358 | 0.3156 | Yes |
| 33 | EHHADH | EHHADH Entrez, Source | enoyl-Coenzyme A, hydratase/3-hydroxyacyl Coenzyme A dehydrogenase | 375 | 0.349 | 0.3208 | Yes |
| 34 | GLYAT | GLYAT Entrez, Source | glycine-N-acyltransferase | 379 | 0.348 | 0.3268 | Yes |
| 35 | DEFB1 | DEFB1 Entrez, Source | defensin, beta 1 | 429 | 0.322 | 0.3288 | Yes |
| 36 | MYH10 | MYH10 Entrez, Source | myosin, heavy chain 10, non-muscle | 442 | 0.313 | 0.3335 | Yes |
| 37 | ALDH3A2 | ALDH3A2 Entrez, Source | aldehyde dehydrogenase 3 family, member A2 | 526 | 0.277 | 0.3320 | Yes |
| 38 | AHCY | AHCY Entrez, Source | S-adenosylhomocysteine hydrolase | 551 | 0.269 | 0.3350 | Yes |
| 39 | ASS1 | ASS1 Entrez, Source | argininosuccinate synthetase 1 | 556 | 0.268 | 0.3395 | Yes |
| 40 | PHYH | PHYH Entrez, Source | phytanoyl-CoA 2-hydroxylase | 566 | 0.265 | 0.3435 | Yes |
| 41 | ALDH5A1 | ALDH5A1 Entrez, Source | aldehyde dehydrogenase 5 family, member A1 (succinate-semialdehyde dehydrogenase) | 595 | 0.258 | 0.3460 | Yes |
| 42 | PPP2R3A | PPP2R3A Entrez, Source | protein phosphatase 2 (formerly 2A), regulatory subunit B'', alpha | 603 | 0.257 | 0.3501 | Yes |
| 43 | ABCD3 | ABCD3 Entrez, Source | ATP-binding cassette, sub-family D (ALD), member 3 | 622 | 0.251 | 0.3532 | Yes |
| 44 | SLC37A4 | SLC37A4 Entrez, Source | solute carrier family 37 (glycerol-6-phosphate transporter), member 4 | 623 | 0.251 | 0.3577 | Yes |
| 45 | ACAA2 | ACAA2 Entrez, Source | acetyl-Coenzyme A acyltransferase 2 (mitochondrial 3-oxoacyl-Coenzyme A thiolase) | 624 | 0.251 | 0.3622 | Yes |
| 46 | BPHL | BPHL Entrez, Source | biphenyl hydrolase-like (serine hydrolase; breast epithelial mucin-associated antigen) | 652 | 0.245 | 0.3645 | Yes |
| 47 | HRSP12 | HRSP12 Entrez, Source | heat-responsive protein 12 | 654 | 0.244 | 0.3687 | Yes |
| 48 | ENPEP | ENPEP Entrez, Source | glutamyl aminopeptidase (aminopeptidase A) | 658 | 0.244 | 0.3729 | Yes |
| 49 | AFM | AFM Entrez, Source | afamin | 681 | 0.239 | 0.3755 | Yes |
| 50 | APOM | APOM Entrez, Source | apolipoprotein M | 687 | 0.238 | 0.3793 | Yes |
| 51 | GJB1 | GJB1 Entrez, Source | gap junction protein, beta 1, 32kDa (connexin 32, Charcot-Marie-Tooth neuropathy, X-linked) | 690 | 0.237 | 0.3834 | Yes |
| 52 | PTPRD | PTPRD Entrez, Source | protein tyrosine phosphatase, receptor type, D | 731 | 0.229 | 0.3844 | Yes |
| 53 | GPC3 | GPC3 Entrez, Source | glypican 3 | 757 | 0.224 | 0.3865 | Yes |
| 54 | PEX3 | PEX3 Entrez, Source | peroxisomal biogenesis factor 3 | 760 | 0.223 | 0.3904 | Yes |
| 55 | GCHFR | GCHFR Entrez, Source | GTP cyclohydrolase I feedback regulator | 762 | 0.223 | 0.3943 | Yes |
| 56 | HPN | HPN Entrez, Source | hepsin (transmembrane protease, serine 1) | 775 | 0.221 | 0.3973 | Yes |
| 57 | RNF14 | RNF14 Entrez, Source | ring finger protein 14 | 836 | 0.207 | 0.3964 | Yes |
| 58 | SERPINA5 | SERPINA5 Entrez, Source | serpin peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 5 | 876 | 0.199 | 0.3970 | Yes |
| 59 | SCP2 | SCP2 Entrez, Source | sterol carrier protein 2 | 890 | 0.197 | 0.3995 | Yes |
| 60 | BCKDHB | BCKDHB Entrez, Source | branched chain keto acid dehydrogenase E1, beta polypeptide (maple syrup urine disease) | 906 | 0.195 | 0.4018 | Yes |
| 61 | ACO2 | ACO2 Entrez, Source | aconitase 2, mitochondrial | 940 | 0.189 | 0.4027 | Yes |
| 62 | CRYZ | CRYZ Entrez, Source | crystallin, zeta (quinone reductase) | 951 | 0.188 | 0.4053 | Yes |
| 63 | KL | KL Entrez, Source | klotho | 971 | 0.186 | 0.4071 | Yes |
| 64 | QPRT | QPRT Entrez, Source | quinolinate phosphoribosyltransferase (nicotinate-nucleotide pyrophosphorylase (carboxylating)) | 1002 | 0.182 | 0.4081 | Yes |
| 65 | BHMT | BHMT Entrez, Source | betaine-homocysteine methyltransferase | 1054 | 0.177 | 0.4073 | Yes |
| 66 | GPSN2 | GPSN2 Entrez, Source | glycoprotein, synaptic 2 | 1058 | 0.175 | 0.4102 | Yes |
| 67 | EPHX2 | EPHX2 Entrez, Source | epoxide hydrolase 2, cytoplasmic | 1108 | 0.170 | 0.4095 | No |
| 68 | PRODH2 | PRODH2 Entrez, Source | proline dehydrogenase (oxidase) 2 | 1197 | 0.161 | 0.4056 | No |
| 69 | TACSTD1 | TACSTD1 Entrez, Source | tumor-associated calcium signal transducer 1 | 1278 | 0.153 | 0.4022 | No |
| 70 | TRIM2 | TRIM2 Entrez, Source | tripartite motif-containing 2 | 1300 | 0.150 | 0.4032 | No |
| 71 | MSH3 | MSH3 Entrez, Source | mutS homolog 3 (E. coli) | 1377 | 0.144 | 0.4000 | No |
| 72 | PRKCA | PRKCA Entrez, Source | protein kinase C, alpha | 1423 | 0.141 | 0.3990 | No |
| 73 | ACAT1 | ACAT1 Entrez, Source | acetyl-Coenzyme A acetyltransferase 1 (acetoacetyl Coenzyme A thiolase) | 1482 | 0.136 | 0.3970 | No |
| 74 | MAOB | MAOB Entrez, Source | monoamine oxidase B | 1531 | 0.133 | 0.3957 | No |
| 75 | PRDX3 | PRDX3 Entrez, Source | peroxiredoxin 3 | 1534 | 0.132 | 0.3979 | No |
| 76 | IGFBP2 | IGFBP2 Entrez, Source | insulin-like growth factor binding protein 2, 36kDa | 1556 | 0.131 | 0.3986 | No |
| 77 | CTBP1 | CTBP1 Entrez, Source | C-terminal binding protein 1 | 1682 | 0.124 | 0.3912 | No |
| 78 | HPGD | HPGD Entrez, Source | hydroxyprostaglandin dehydrogenase 15-(NAD) | 1689 | 0.123 | 0.3930 | No |
| 79 | PEX7 | PEX7 Entrez, Source | peroxisomal biogenesis factor 7 | 1696 | 0.123 | 0.3947 | No |
| 80 | ACADSB | ACADSB Entrez, Source | acyl-Coenzyme A dehydrogenase, short/branched chain | 1715 | 0.122 | 0.3955 | No |
| 81 | CBR4 | CBR4 Entrez, Source | carbonyl reductase 4 | 1733 | 0.121 | 0.3964 | No |
| 82 | COX7B | COX7B Entrez, Source | cytochrome c oxidase subunit VIIb | 1747 | 0.119 | 0.3975 | No |
| 83 | MINPP1 | MINPP1 Entrez, Source | multiple inositol polyphosphate histidine phosphatase, 1 | 1760 | 0.119 | 0.3987 | No |
| 84 | CRYM | CRYM Entrez, Source | crystallin, mu | 1874 | 0.113 | 0.3920 | No |
| 85 | TNFRSF11B | TNFRSF11B Entrez, Source | tumor necrosis factor receptor superfamily, member 11b (osteoprotegerin) | 1914 | 0.111 | 0.3910 | No |
| 86 | DZIP1 | DZIP1 Entrez, Source | DAZ interacting protein 1 | 1960 | 0.109 | 0.3895 | No |
| 87 | TST | TST Entrez, Source | thiosulfate sulfurtransferase (rhodanese) | 1969 | 0.109 | 0.3908 | No |
| 88 | APOH | APOH Entrez, Source | apolipoprotein H (beta-2-glycoprotein I) | 2011 | 0.107 | 0.3896 | No |
| 89 | RNASE4 | RNASE4 Entrez, Source | ribonuclease, RNase A family, 4 | 2064 | 0.105 | 0.3875 | No |
| 90 | CTH | CTH Entrez, Source | cystathionase (cystathionine gamma-lyase) | 2088 | 0.104 | 0.3875 | No |
| 91 | ASPA | ASPA Entrez, Source | aspartoacylase (Canavan disease) | 2090 | 0.104 | 0.3893 | No |
| 92 | CLINT1 | CLINT1 Entrez, Source | clathrin interactor 1 | 2120 | 0.103 | 0.3889 | No |
| 93 | ALDOB | ALDOB Entrez, Source | aldolase B, fructose-bisphosphate | 2152 | 0.101 | 0.3884 | No |
| 94 | MAOA | MAOA Entrez, Source | monoamine oxidase A | 2159 | 0.101 | 0.3897 | No |
| 95 | PRKRA | PRKRA Entrez, Source | protein kinase, interferon-inducible double stranded RNA dependent activator | 2179 | 0.100 | 0.3900 | No |
| 96 | COMT | COMT Entrez, Source | catechol-O-methyltransferase | 2208 | 0.099 | 0.3897 | No |
| 97 | ABAT | ABAT Entrez, Source | 4-aminobutyrate aminotransferase | 2398 | 0.092 | 0.3768 | No |
| 98 | DDT | DDT Entrez, Source | D-dopachrome tautomerase | 2453 | 0.090 | 0.3742 | No |
| 99 | CLTB | CLTB Entrez, Source | clathrin, light chain (Lcb) | 2459 | 0.089 | 0.3754 | No |
| 100 | PDZK1IP1 | PDZK1IP1 Entrez, Source | PDZK1 interacting protein 1 | 2512 | 0.087 | 0.3730 | No |
| 101 | CA4 | CA4 Entrez, Source | carbonic anhydrase IV | 2514 | 0.087 | 0.3745 | No |
| 102 | FBXO9 | FBXO9 Entrez, Source | F-box protein 9 | 2539 | 0.086 | 0.3742 | No |
| 103 | ALDH2 | ALDH2 Entrez, Source | aldehyde dehydrogenase 2 family (mitochondrial) | 2544 | 0.086 | 0.3754 | No |
| 104 | ATP5O | ATP5O Entrez, Source | ATP synthase, H+ transporting, mitochondrial F1 complex, O subunit (oligomycin sensitivity conferring protein) | 2593 | 0.084 | 0.3732 | No |
| 105 | FYN | FYN Entrez, Source | FYN oncogene related to SRC, FGR, YES | 2623 | 0.083 | 0.3724 | No |
| 106 | APLP2 | APLP2 Entrez, Source | amyloid beta (A4) precursor-like protein 2 | 2657 | 0.082 | 0.3714 | No |
| 107 | MDH1 | MDH1 Entrez, Source | malate dehydrogenase 1, NAD (soluble) | 2674 | 0.081 | 0.3716 | No |
| 108 | DDC | DDC Entrez, Source | dopa decarboxylase (aromatic L-amino acid decarboxylase) | 2692 | 0.081 | 0.3717 | No |
| 109 | ZMYND11 | ZMYND11 Entrez, Source | zinc finger, MYND domain containing 11 | 2706 | 0.080 | 0.3721 | No |
| 110 | ATP5J | ATP5J Entrez, Source | ATP synthase, H+ transporting, mitochondrial F0 complex, subunit F6 | 2738 | 0.079 | 0.3712 | No |
| 111 | CDH1 | CDH1 Entrez, Source | cadherin 1, type 1, E-cadherin (epithelial) | 2848 | 0.076 | 0.3641 | No |
| 112 | ALDH9A1 | ALDH9A1 Entrez, Source | aldehyde dehydrogenase 9 family, member A1 | 2893 | 0.074 | 0.3621 | No |
| 113 | ACOX1 | ACOX1 Entrez, Source | acyl-Coenzyme A oxidase 1, palmitoyl | 2947 | 0.073 | 0.3593 | No |
| 114 | ENPP2 | ENPP2 Entrez, Source | ectonucleotide pyrophosphatase/phosphodiesterase 2 (autotaxin) | 2952 | 0.072 | 0.3603 | No |
| 115 | HNMT | HNMT Entrez, Source | histamine N-methyltransferase | 2986 | 0.072 | 0.3590 | No |
| 116 | HADH | HADH Entrez, Source | hydroxyacyl-Coenzyme A dehydrogenase | 3083 | 0.069 | 0.3528 | No |
| 117 | FPGT | FPGT Entrez, Source | fucose-1-phosphate guanylyltransferase | 3102 | 0.068 | 0.3527 | No |
| 118 | IDH1 | IDH1 Entrez, Source | isocitrate dehydrogenase 1 (NADP+), soluble | 3161 | 0.067 | 0.3494 | No |
| 119 | RAB7L1 | RAB7L1 Entrez, Source | RAB7, member RAS oncogene family-like 1 | 3207 | 0.066 | 0.3471 | No |
| 120 | ETFA | ETFA Entrez, Source | electron-transfer-flavoprotein, alpha polypeptide (glutaric aciduria II) | 3209 | 0.065 | 0.3482 | No |
| 121 | CYCS | CYCS Entrez, Source | cytochrome c, somatic | 3256 | 0.064 | 0.3458 | No |
| 122 | SDHC | SDHC Entrez, Source | succinate dehydrogenase complex, subunit C, integral membrane protein, 15kDa | 3311 | 0.062 | 0.3428 | No |
| 123 | COL4A4 | COL4A4 Entrez, Source | collagen, type IV, alpha 4 | 3320 | 0.062 | 0.3433 | No |
| 124 | COX6A1 | COX6A1 Entrez, Source | cytochrome c oxidase subunit VIa polypeptide 1 | 3335 | 0.062 | 0.3433 | No |
| 125 | PPM1B | PPM1B Entrez, Source | protein phosphatase 1B (formerly 2C), magnesium-dependent, beta isoform | 3422 | 0.060 | 0.3377 | No |
| 126 | NR1D2 | NR1D2 Entrez, Source | nuclear receptor subfamily 1, group D, member 2 | 3431 | 0.060 | 0.3382 | No |
| 127 | AKR7A2 | AKR7A2 Entrez, Source | aldo-keto reductase family 7, member A2 (aflatoxin aldehyde reductase) | 3458 | 0.059 | 0.3372 | No |
| 128 | ACO1 | ACO1 Entrez, Source | aconitase 1, soluble | 3462 | 0.059 | 0.3381 | No |
| 129 | DNM1L | DNM1L Entrez, Source | dynamin 1-like | 3499 | 0.058 | 0.3363 | No |
| 130 | HAGH | HAGH Entrez, Source | hydroxyacylglutathione hydrolase | 3639 | 0.054 | 0.3266 | No |
| 131 | SLC22A2 | SLC22A2 Entrez, Source | solute carrier family 22 (organic cation transporter), member 2 | 3656 | 0.053 | 0.3263 | No |
| 132 | SEPHS2 | SEPHS2 Entrez, Source | selenophosphate synthetase 2 | 3685 | 0.053 | 0.3251 | No |
| 133 | ABP1 | ABP1 Entrez, Source | amiloride binding protein 1 (amine oxidase (copper-containing)) | 3774 | 0.051 | 0.3192 | No |
| 134 | SMPDL3A | SMPDL3A Entrez, Source | sphingomyelin phosphodiesterase, acid-like 3A | 3776 | 0.051 | 0.3200 | No |
| 135 | ACADM | ACADM Entrez, Source | acyl-Coenzyme A dehydrogenase, C-4 to C-12 straight chain | 3794 | 0.050 | 0.3196 | No |
| 136 | SLC1A1 | SLC1A1 Entrez, Source | solute carrier family 1 (neuronal/epithelial high affinity glutamate transporter, system Xag), member 1 | 3801 | 0.050 | 0.3201 | No |
| 137 | CALB1 | CALB1 Entrez, Source | calbindin 1, 28kDa | 3847 | 0.049 | 0.3175 | No |
| 138 | IGF1R | IGF1R Entrez, Source | insulin-like growth factor 1 receptor | 3890 | 0.048 | 0.3151 | No |
| 139 | ERBB3 | ERBB3 Entrez, Source | v-erb-b2 erythroblastic leukemia viral oncogene homolog 3 (avian) | 4005 | 0.044 | 0.3071 | No |
| 140 | BAMBI | BAMBI Entrez, Source | BMP and activin membrane-bound inhibitor homolog (Xenopus laevis) | 4035 | 0.044 | 0.3057 | No |
| 141 | PRDX2 | PRDX2 Entrez, Source | peroxiredoxin 2 | 4063 | 0.043 | 0.3044 | No |
| 142 | PDE8A | PDE8A Entrez, Source | phosphodiesterase 8A | 4161 | 0.041 | 0.2976 | No |
| 143 | SULT1C1 | SULT1C1 Entrez, Source | sulfotransferase family, cytosolic, 1C, member 1 | 4188 | 0.040 | 0.2963 | No |
| 144 | DSTN | DSTN Entrez, Source | destrin (actin depolymerizing factor) | 4198 | 0.040 | 0.2963 | No |
| 145 | G3BP | G3BP Entrez, Source | - | 4201 | 0.039 | 0.2969 | No |
| 146 | SDHA | SDHA Entrez, Source | succinate dehydrogenase complex, subunit A, flavoprotein (Fp) | 4257 | 0.038 | 0.2933 | No |
| 147 | HSD11B2 | HSD11B2 Entrez, Source | hydroxysteroid (11-beta) dehydrogenase 2 | 4357 | 0.037 | 0.2864 | No |
| 148 | CLDN8 | CLDN8 Entrez, Source | claudin 8 | 4382 | 0.036 | 0.2852 | No |
| 149 | ESRRG | ESRRG Entrez, Source | estrogen-related receptor gamma | 4402 | 0.036 | 0.2844 | No |
| 150 | KDR | KDR Entrez, Source | kinase insert domain receptor (a type III receptor tyrosine kinase) | 4442 | 0.035 | 0.2820 | No |
| 151 | CGRRF1 | CGRRF1 Entrez, Source | cell growth regulator with ring finger domain 1 | 4445 | 0.035 | 0.2824 | No |
| 152 | SLC25A1 | SLC25A1 Entrez, Source | solute carrier family 25 (mitochondrial carrier; citrate transporter), member 1 | 4469 | 0.035 | 0.2813 | No |
| 153 | KCNJ15 | KCNJ15 Entrez, Source | potassium inwardly-rectifying channel, subfamily J, member 15 | 4470 | 0.035 | 0.2819 | No |
| 154 | IGFBP4 | IGFBP4 Entrez, Source | insulin-like growth factor binding protein 4 | 4578 | 0.032 | 0.2742 | No |
| 155 | ADH6 | ADH6 Entrez, Source | alcohol dehydrogenase 6 (class V) | 4624 | 0.032 | 0.2714 | No |
| 156 | SC5DL | SC5DL Entrez, Source | sterol-C5-desaturase (ERG3 delta-5-desaturase homolog, fungal)-like | 4644 | 0.031 | 0.2704 | No |
| 157 | PTPN3 | PTPN3 Entrez, Source | protein tyrosine phosphatase, non-receptor type 3 | 4694 | 0.030 | 0.2672 | No |
| 158 | PRKAR1A | PRKAR1A Entrez, Source | protein kinase, cAMP-dependent, regulatory, type I, alpha (tissue specific extinguisher 1) | 4750 | 0.029 | 0.2635 | No |
| 159 | NDUFS4 | NDUFS4 Entrez, Source | NADH dehydrogenase (ubiquinone) Fe-S protein 4, 18kDa (NADH-coenzyme Q reductase) | 4767 | 0.029 | 0.2628 | No |
| 160 | UGT8 | UGT8 Entrez, Source | UDP glycosyltransferase 8 (UDP-galactose ceramide galactosyltransferase) | 4812 | 0.028 | 0.2599 | No |
| 161 | ATP5A1 | ATP5A1 Entrez, Source | ATP synthase, H+ transporting, mitochondrial F1 complex, alpha subunit 1, cardiac muscle | 4884 | 0.026 | 0.2549 | No |
| 162 | PPP2CB | PPP2CB Entrez, Source | protein phosphatase 2 (formerly 2A), catalytic subunit, beta isoform | 5055 | 0.023 | 0.2422 | No |
| 163 | SLC12A3 | SLC12A3 Entrez, Source | solute carrier family 12 (sodium/chloride transporters), member 3 | 5080 | 0.023 | 0.2408 | No |
| 164 | PDCD8 | PDCD8 Entrez, Source | programmed cell death 8 (apoptosis-inducing factor) | 5123 | 0.022 | 0.2379 | No |
| 165 | SLC26A4 | SLC26A4 Entrez, Source | solute carrier family 26, member 4 | 5361 | 0.018 | 0.2200 | No |
| 166 | FGF9 | FGF9 Entrez, Source | fibroblast growth factor 9 (glia-activating factor) | 5381 | 0.017 | 0.2188 | No |
| 167 | FAM3C | FAM3C Entrez, Source | family with sequence similarity 3, member C | 5436 | 0.016 | 0.2149 | No |
| 168 | UQCRC2 | UQCRC2 Entrez, Source | ubiquinol-cytochrome c reductase core protein II | 5487 | 0.015 | 0.2114 | No |
| 169 | MXI1 | MXI1 Entrez, Source | MAX interactor 1 | 5523 | 0.014 | 0.2089 | No |
| 170 | SERPINI1 | SERPINI1 Entrez, Source | serpin peptidase inhibitor, clade I (neuroserpin), member 1 | 5563 | 0.013 | 0.2062 | No |
| 171 | LRPAP1 | LRPAP1 Entrez, Source | low density lipoprotein receptor-related protein associated protein 1 | 5727 | 0.010 | 0.1938 | No |
| 172 | IMMT | IMMT Entrez, Source | inner membrane protein, mitochondrial (mitofilin) | 5761 | 0.010 | 0.1914 | No |
| 173 | ADD3 | ADD3 Entrez, Source | adducin 3 (gamma) | 5820 | 0.009 | 0.1871 | No |
| 174 | SLC7A8 | SLC7A8 Entrez, Source | solute carrier family 7 (cationic amino acid transporter, y+ system), member 8 | 5837 | 0.008 | 0.1860 | No |
| 175 | SC4MOL | SC4MOL Entrez, Source | sterol-C4-methyl oxidase-like | 5880 | 0.008 | 0.1829 | No |
| 176 | UQCRC1 | UQCRC1 Entrez, Source | ubiquinol-cytochrome c reductase core protein I | 5927 | 0.007 | 0.1795 | No |
| 177 | USP14 | USP14 Entrez, Source | ubiquitin specific peptidase 14 (tRNA-guanine transglycosylase) | 5928 | 0.007 | 0.1796 | No |
| 178 | PEX11B | PEX11B Entrez, Source | peroxisomal biogenesis factor 11B | 5988 | 0.005 | 0.1752 | No |
| 179 | SCAMP1 | SCAMP1 Entrez, Source | secretory carrier membrane protein 1 | 6004 | 0.005 | 0.1741 | No |
| 180 | NDUFA5 | NDUFA5 Entrez, Source | NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 5, 13kDa | 6009 | 0.005 | 0.1739 | No |
| 181 | UQCRFS1 | UQCRFS1 Entrez, Source | ubiquinol-cytochrome c reductase, Rieske iron-sulfur polypeptide 1 | 6010 | 0.005 | 0.1740 | No |
| 182 | DNAJB9 | DNAJB9 Entrez, Source | DnaJ (Hsp40) homolog, subfamily B, member 9 | 6082 | 0.004 | 0.1686 | No |
| 183 | DLD | DLD Entrez, Source | dihydrolipoamide dehydrogenase | 6088 | 0.003 | 0.1683 | No |
| 184 | FGF1 | FGF1 Entrez, Source | fibroblast growth factor 1 (acidic) | 6094 | 0.003 | 0.1679 | No |
| 185 | SLC22A6 | SLC22A6 Entrez, Source | solute carrier family 22 (organic anion transporter), member 6 | 6230 | 0.001 | 0.1576 | No |
| 186 | PTHR1 | PTHR1 Entrez, Source | parathyroid hormone receptor 1 | 6271 | 0.000 | 0.1545 | No |
| 187 | ALB | ALB Entrez, Source | albumin | 6275 | 0.000 | 0.1543 | No |
| 188 | PFN2 | PFN2 Entrez, Source | profilin 2 | 6281 | 0.000 | 0.1539 | No |
| 189 | NCKAP1 | NCKAP1 Entrez, Source | NCK-associated protein 1 | 6284 | 0.000 | 0.1537 | No |
| 190 | DPP4 | DPP4 Entrez, Source | dipeptidyl-peptidase 4 (CD26, adenosine deaminase complexing protein 2) | 6342 | -0.001 | 0.1493 | No |
| 191 | MFAP3L | MFAP3L Entrez, Source | microfibrillar-associated protein 3-like | 6343 | -0.001 | 0.1494 | No |
| 192 | IGFBP5 | IGFBP5 Entrez, Source | insulin-like growth factor binding protein 5 | 6450 | -0.003 | 0.1412 | No |
| 193 | HPD | HPD Entrez, Source | 4-hydroxyphenylpyruvate dioxygenase | 6481 | -0.003 | 0.1390 | No |
| 194 | CAP2 | CAP2 Entrez, Source | CAP, adenylate cyclase-associated protein, 2 (yeast) | 6484 | -0.003 | 0.1389 | No |
| 195 | HSPD1 | HSPD1 Entrez, Source | heat shock 60kDa protein 1 (chaperonin) | 6540 | -0.004 | 0.1347 | No |
| 196 | ANK3 | ANK3 Entrez, Source | ankyrin 3, node of Ranvier (ankyrin G) | 6582 | -0.005 | 0.1317 | No |
| 197 | RAB1A | RAB1A Entrez, Source | RAB1A, member RAS oncogene family | 6729 | -0.008 | 0.1206 | No |
| 198 | PCBD1 | PCBD1 Entrez, Source | pterin-4 alpha-carbinolamine dehydratase/dimerization cofactor of hepatocyte nuclear factor 1 alpha (TCF1) | 6764 | -0.008 | 0.1181 | No |
| 199 | TOB1 | TOB1 Entrez, Source | transducer of ERBB2, 1 | 6785 | -0.009 | 0.1167 | No |
| 200 | ACY1 | ACY1 Entrez, Source | aminoacylase 1 | 6868 | -0.010 | 0.1106 | No |
| 201 | FABP1 | FABP1 Entrez, Source | fatty acid binding protein 1, liver | 6898 | -0.010 | 0.1085 | No |
| 202 | ITGA6 | ITGA6 Entrez, Source | integrin, alpha 6 | 7051 | -0.013 | 0.0971 | No |
| 203 | HMGCL | HMGCL Entrez, Source | 3-hydroxymethyl-3-methylglutaryl-Coenzyme A lyase (hydroxymethylglutaricaciduria) | 7083 | -0.014 | 0.0949 | No |
| 204 | OGG1 | OGG1 Entrez, Source | 8-oxoguanine DNA glycosylase | 7089 | -0.014 | 0.0948 | No |
| 205 | SOD1 | SOD1 Entrez, Source | superoxide dismutase 1, soluble (amyotrophic lateral sclerosis 1 (adult)) | 7093 | -0.014 | 0.0948 | No |
| 206 | AQP1 | AQP1 Entrez, Source | aquaporin 1 (Colton blood group) | 7239 | -0.017 | 0.0839 | No |
| 207 | CRYAA | CRYAA Entrez, Source | crystallin, alpha A | 7303 | -0.018 | 0.0794 | No |
| 208 | DSP | DSP Entrez, Source | desmoplakin | 7304 | -0.018 | 0.0797 | No |
| 209 | PEX19 | PEX19 Entrez, Source | peroxisomal biogenesis factor 19 | 7306 | -0.018 | 0.0800 | No |
| 210 | PHKB | PHKB Entrez, Source | phosphorylase kinase, beta | 7355 | -0.019 | 0.0766 | No |
| 211 | ZNF148 | ZNF148 Entrez, Source | zinc finger protein 148 | 7364 | -0.019 | 0.0763 | No |
| 212 | GPR116 | GPR116 Entrez, Source | G protein-coupled receptor 116 | 7368 | -0.019 | 0.0764 | No |
| 213 | CTSL | CTSL Entrez, Source | cathepsin L | 7390 | -0.020 | 0.0752 | No |
| 214 | QDPR | QDPR Entrez, Source | quinoid dihydropteridine reductase | 7441 | -0.021 | 0.0717 | No |
| 215 | TMED5 | TMED5 Entrez, Source | transmembrane emp24 protein transport domain containing 5 | 7449 | -0.021 | 0.0715 | No |
| 216 | CREB3L2 | CREB3L2 Entrez, Source | cAMP responsive element binding protein 3-like 2 | 7503 | -0.022 | 0.0678 | No |
| 217 | PRKCI | PRKCI Entrez, Source | protein kinase C, iota | 7723 | -0.026 | 0.0514 | No |
| 218 | PHB | PHB Entrez, Source | prohibitin | 7736 | -0.026 | 0.0510 | No |
| 219 | SHMT1 | SHMT1 Entrez, Source | serine hydroxymethyltransferase 1 (soluble) | 7827 | -0.028 | 0.0445 | No |
| 220 | DSCR1 | DSCR1 Entrez, Source | Down syndrome critical region gene 1 | 7972 | -0.031 | 0.0340 | No |
| 221 | HADHB | HADHB Entrez, Source | hydroxyacyl-Coenzyme A dehydrogenase/3-ketoacyl-Coenzyme A thiolase/enoyl-Coenzyme A hydratase (trifunctional protein), beta subunit | 7978 | -0.031 | 0.0341 | No |
| 222 | PARG | PARG Entrez, Source | poly (ADP-ribose) glycohydrolase | 7997 | -0.031 | 0.0333 | No |
| 223 | SMAD4 | SMAD4 Entrez, Source | SMAD, mothers against DPP homolog 4 (Drosophila) | 8001 | -0.031 | 0.0336 | No |
| 224 | CDC42BPA | CDC42BPA Entrez, Source | CDC42 binding protein kinase alpha (DMPK-like) | 8008 | -0.031 | 0.0337 | No |
| 225 | SERINC1 | SERINC1 Entrez, Source | serine incorporator 1 | 8027 | -0.032 | 0.0329 | No |
| 226 | PGM1 | PGM1 Entrez, Source | phosphoglucomutase 1 | 8051 | -0.032 | 0.0317 | No |
| 227 | AP3D1 | AP3D1 Entrez, Source | adaptor-related protein complex 3, delta 1 subunit | 8055 | -0.032 | 0.0321 | No |
| 228 | CYP4F2 | CYP4F2 Entrez, Source | cytochrome P450, family 4, subfamily F, polypeptide 2 | 8129 | -0.034 | 0.0271 | No |
| 229 | HIBCH | HIBCH Entrez, Source | 3-hydroxyisobutyryl-Coenzyme A hydrolase | 8146 | -0.034 | 0.0264 | No |
| 230 | TMED10 | TMED10 Entrez, Source | transmembrane emp24-like trafficking protein 10 (yeast) | 8177 | -0.035 | 0.0248 | No |
| 231 | NDUFS1 | NDUFS1 Entrez, Source | NADH dehydrogenase (ubiquinone) Fe-S protein 1, 75kDa (NADH-coenzyme Q reductase) | 8263 | -0.037 | 0.0189 | No |
| 232 | DAP | DAP Entrez, Source | death-associated protein | 8268 | -0.037 | 0.0192 | No |
| 233 | MYH11 | MYH11 Entrez, Source | myosin, heavy chain 11, smooth muscle | 8278 | -0.037 | 0.0192 | No |
| 234 | RRAGA | RRAGA Entrez, Source | Ras-related GTP binding A | 8307 | -0.037 | 0.0177 | No |
| 235 | NQO2 | NQO2 Entrez, Source | NAD(P)H dehydrogenase, quinone 2 | 8329 | -0.038 | 0.0168 | No |
| 236 | STRN3 | STRN3 Entrez, Source | striatin, calmodulin binding protein 3 | 8342 | -0.038 | 0.0165 | No |
| 237 | WWP1 | WWP1 Entrez, Source | WW domain containing E3 ubiquitin protein ligase 1 | 8375 | -0.039 | 0.0148 | No |
| 238 | VDAC2 | VDAC2 Entrez, Source | voltage-dependent anion channel 2 | 8408 | -0.040 | 0.0130 | No |
| 239 | PLS3 | PLS3 Entrez, Source | plastin 3 (T isoform) | 8424 | -0.040 | 0.0126 | No |
| 240 | PPP2R1A | PPP2R1A Entrez, Source | protein phosphatase 2 (formerly 2A), regulatory subunit A (PR 65), alpha isoform | 8432 | -0.040 | 0.0127 | No |
| 241 | CYB5R3 | CYB5R3 Entrez, Source | cytochrome b5 reductase 3 | 8531 | -0.042 | 0.0059 | No |
| 242 | EFHA1 | EFHA1 Entrez, Source | EF-hand domain family, member A1 | 8541 | -0.042 | 0.0060 | No |
| 243 | ATP2A2 | ATP2A2 Entrez, Source | ATPase, Ca++ transporting, cardiac muscle, slow twitch 2 | 8616 | -0.044 | 0.0011 | No |
| 244 | SLC31A1 | SLC31A1 Entrez, Source | solute carrier family 31 (copper transporters), member 1 | 8681 | -0.045 | -0.0030 | No |
| 245 | YWHAE | YWHAE Entrez, Source | tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, epsilon polypeptide | 8704 | -0.045 | -0.0039 | No |
| 246 | ITM2B | ITM2B Entrez, Source | integral membrane protein 2B | 8720 | -0.046 | -0.0042 | No |
| 247 | HBLD2 | HBLD2 Entrez, Source | HESB like domain containing 2 | 8732 | -0.046 | -0.0043 | No |
| 248 | PJA2 | PJA2 Entrez, Source | praja 2, RING-H2 motif containing | 8736 | -0.046 | -0.0037 | No |
| 249 | CAT | CAT Entrez, Source | catalase | 8873 | -0.049 | -0.0133 | No |
| 250 | AKR1A1 | AKR1A1 Entrez, Source | aldo-keto reductase family 1, member A1 (aldehyde reductase) | 8947 | -0.051 | -0.0180 | No |
| 251 | LRP2 | LRP2 Entrez, Source | low density lipoprotein-related protein 2 | 8982 | -0.051 | -0.0197 | No |
| 252 | PGRMC2 | PGRMC2 Entrez, Source | progesterone receptor membrane component 2 | 9059 | -0.053 | -0.0246 | No |
| 253 | PMPCB | PMPCB Entrez, Source | peptidase (mitochondrial processing) beta | 9068 | -0.053 | -0.0242 | No |
| 254 | GLS | GLS Entrez, Source | glutaminase | 9073 | -0.053 | -0.0236 | No |
| 255 | SHANK2 | SHANK2 Entrez, Source | SH3 and multiple ankyrin repeat domains 2 | 9084 | -0.054 | -0.0234 | No |
| 256 | PLG | PLG Entrez, Source | plasminogen | 9113 | -0.054 | -0.0246 | No |
| 257 | TEGT | TEGT Entrez, Source | testis enhanced gene transcript (BAX inhibitor 1) | 9169 | -0.055 | -0.0278 | No |
| 258 | TOMM20 | TOMM20 Entrez, Source | translocase of outer mitochondrial membrane 20 homolog (yeast) | 9181 | -0.056 | -0.0277 | No |
| 259 | ANKRD46 | ANKRD46 Entrez, Source | ankyrin repeat domain 46 | 9239 | -0.057 | -0.0311 | No |
| 260 | GHITM | GHITM Entrez, Source | growth hormone inducible transmembrane protein | 9256 | -0.057 | -0.0313 | No |
| 261 | HSPE1 | HSPE1 Entrez, Source | heat shock 10kDa protein 1 (chaperonin 10) | 9296 | -0.058 | -0.0332 | No |
| 262 | PGRMC1 | PGRMC1 Entrez, Source | progesterone receptor membrane component 1 | 9378 | -0.060 | -0.0384 | No |
| 263 | HSPA8 | HSPA8 Entrez, Source | heat shock 70kDa protein 8 | 9449 | -0.062 | -0.0427 | No |
| 264 | GULP1 | GULP1 Entrez, Source | GULP, engulfment adaptor PTB domain containing 1 | 9461 | -0.062 | -0.0424 | No |
| 265 | PODXL | PODXL Entrez, Source | podocalyxin-like | 9478 | -0.062 | -0.0425 | No |
| 266 | AQP3 | AQP3 Entrez, Source | aquaporin 3 (Gill blood group) | 9501 | -0.063 | -0.0431 | No |
| 267 | DDX3X | DDX3X Entrez, Source | DEAD (Asp-Glu-Ala-Asp) box polypeptide 3, X-linked | 9594 | -0.065 | -0.0490 | No |
| 268 | VDAC1 | VDAC1 Entrez, Source | voltage-dependent anion channel 1 | 9595 | -0.065 | -0.0479 | No |
| 269 | COPS5 | COPS5 Entrez, Source | COP9 constitutive photomorphogenic homolog subunit 5 (Arabidopsis) | 9599 | -0.065 | -0.0469 | No |
| 270 | STCH | STCH Entrez, Source | stress 70 protein chaperone, microsome-associated, 60kDa | 9623 | -0.066 | -0.0475 | No |
| 271 | HTRA1 | HTRA1 Entrez, Source | HtrA serine peptidase 1 | 9698 | -0.068 | -0.0520 | No |
| 272 | TSPYL4 | TSPYL4 Entrez, Source | TSPY-like 4 | 9720 | -0.069 | -0.0524 | No |
| 273 | PEBP1 | PEBP1 Entrez, Source | phosphatidylethanolamine binding protein 1 | 9842 | -0.072 | -0.0604 | No |
| 274 | BNIP3 | BNIP3 Entrez, Source | BCL2/adenovirus E1B 19kDa interacting protein 3 | 9855 | -0.073 | -0.0600 | No |
| 275 | ABCC2 | ABCC2 Entrez, Source | ATP-binding cassette, sub-family C (CFTR/MRP), member 2 | 9965 | -0.075 | -0.0671 | No |
| 276 | MORF4L2 | MORF4L2 Entrez, Source | mortality factor 4 like 2 | 10054 | -0.078 | -0.0725 | No |
| 277 | COPS2 | COPS2 Entrez, Source | COP9 constitutive photomorphogenic homolog subunit 2 (Arabidopsis) | 10059 | -0.078 | -0.0714 | No |
| 278 | CDKN1C | CDKN1C Entrez, Source | cyclin-dependent kinase inhibitor 1C (p57, Kip2) | 10104 | -0.079 | -0.0733 | No |
| 279 | RAB2 | RAB2 Entrez, Source | RAB2, member RAS oncogene family | 10106 | -0.079 | -0.0720 | No |
| 280 | RFK | RFK Entrez, Source | riboflavin kinase | 10120 | -0.080 | -0.0716 | No |
| 281 | SNRPN /// SNURF | 10197 | -0.082 | -0.0760 | No | ||
| 282 | COPA | COPA Entrez, Source | coatomer protein complex, subunit alpha | 10279 | -0.084 | -0.0807 | No |
| 283 | RDX | RDX Entrez, Source | radixin | 10400 | -0.087 | -0.0884 | No |
| 284 | GRB14 | GRB14 Entrez, Source | growth factor receptor-bound protein 14 | 10443 | -0.089 | -0.0900 | No |
| 285 | CALM1 | CALM1 Entrez, Source | calmodulin 1 (phosphorylase kinase, delta) | 10674 | -0.096 | -0.1060 | No |
| 286 | CA2 | CA2 Entrez, Source | carbonic anhydrase II | 10759 | -0.099 | -0.1107 | No |
| 287 | GATM | GATM Entrez, Source | glycine amidinotransferase (L-arginine:glycine amidinotransferase) | 10782 | -0.100 | -0.1106 | No |
| 288 | NEK4 | NEK4 Entrez, Source | NIMA (never in mitosis gene a)-related kinase 4 | 10801 | -0.101 | -0.1102 | No |
| 289 | FOLH1 | FOLH1 Entrez, Source | folate hydrolase (prostate-specific membrane antigen) 1 | 10825 | -0.102 | -0.1101 | No |
| 290 | ANPEP | ANPEP Entrez, Source | alanyl (membrane) aminopeptidase (aminopeptidase N, aminopeptidase M, microsomal aminopeptidase, CD13, p150) | 10917 | -0.106 | -0.1153 | No |
| 291 | C1QBP | C1QBP Entrez, Source | complement component 1, q subcomponent binding protein | 10931 | -0.106 | -0.1144 | No |
| 292 | ANXA7 | ANXA7 Entrez, Source | annexin A7 | 10981 | -0.108 | -0.1162 | No |
| 293 | NR3C2 | NR3C2 Entrez, Source | nuclear receptor subfamily 3, group C, member 2 | 11091 | -0.112 | -0.1226 | No |
| 294 | SORD | SORD Entrez, Source | sorbitol dehydrogenase | 11093 | -0.112 | -0.1207 | No |
| 295 | LRPPRC | LRPPRC Entrez, Source | leucine-rich PPR-motif containing | 11099 | -0.112 | -0.1191 | No |
| 296 | HRASLS3 | HRASLS3 Entrez, Source | HRAS-like suppressor 3 | 11191 | -0.116 | -0.1240 | No |
| 297 | RABGGTB | RABGGTB Entrez, Source | Rab geranylgeranyltransferase, beta subunit | 11200 | -0.116 | -0.1225 | No |
| 298 | STRAP | STRAP Entrez, Source | serine/threonine kinase receptor associated protein | 11237 | -0.117 | -0.1232 | No |
| 299 | NARS | NARS Entrez, Source | asparaginyl-tRNA synthetase | 11247 | -0.118 | -0.1218 | No |
| 300 | KHK | KHK Entrez, Source | ketohexokinase (fructokinase) | 11294 | -0.120 | -0.1232 | No |
| 301 | PIK3R1 | PIK3R1 Entrez, Source | phosphoinositide-3-kinase, regulatory subunit 1 (p85 alpha) | 11304 | -0.121 | -0.1217 | No |
| 302 | MPDZ | MPDZ Entrez, Source | multiple PDZ domain protein | 11344 | -0.122 | -0.1225 | No |
| 303 | ACPP | ACPP Entrez, Source | acid phosphatase, prostate | 11357 | -0.123 | -0.1213 | No |
| 304 | SERBP1 | SERBP1 Entrez, Source | SERPINE1 mRNA binding protein 1 | 11399 | -0.125 | -0.1222 | No |
| 305 | PCBP2 | PCBP2 Entrez, Source | poly(rC) binding protein 2 | 11406 | -0.125 | -0.1204 | No |
| 306 | IMPA2 | IMPA2 Entrez, Source | inositol(myo)-1(or 4)-monophosphatase 2 | 11426 | -0.126 | -0.1196 | No |
| 307 | MRPS18B | MRPS18B Entrez, Source | mitochondrial ribosomal protein S18B | 11505 | -0.129 | -0.1233 | No |
| 308 | RTN4 | RTN4 Entrez, Source | reticulon 4 | 11618 | -0.135 | -0.1295 | No |
| 309 | CFDP1 | CFDP1 Entrez, Source | craniofacial development protein 1 | 11738 | -0.140 | -0.1362 | No |
| 310 | TSC22D1 | TSC22D1 Entrez, Source | TSC22 domain family, member 1 | 11757 | -0.142 | -0.1350 | No |
| 311 | OAT | OAT Entrez, Source | ornithine aminotransferase (gyrate atrophy) | 11781 | -0.143 | -0.1342 | No |
| 312 | CLASP2 | CLASP2 Entrez, Source | cytoplasmic linker associated protein 2 | 11792 | -0.144 | -0.1324 | No |
| 313 | ANKRD15 | ANKRD15 Entrez, Source | ankyrin repeat domain 15 | 11802 | -0.145 | -0.1305 | No |
| 314 | PAFAH1B1 | PAFAH1B1 Entrez, Source | platelet-activating factor acetylhydrolase, isoform Ib, alpha subunit 45kDa | 11805 | -0.145 | -0.1281 | No |
| 315 | PTGER3 | PTGER3 Entrez, Source | prostaglandin E receptor 3 (subtype EP3) | 11830 | -0.146 | -0.1273 | No |
| 316 | SLC7A7 | SLC7A7 Entrez, Source | solute carrier family 7 (cationic amino acid transporter, y+ system), member 7 | 11894 | -0.150 | -0.1295 | No |
| 317 | HDGF | HDGF Entrez, Source | hepatoma-derived growth factor (high-mobility group protein 1-like) | 11910 | -0.150 | -0.1280 | No |
| 318 | WEE1 | WEE1 Entrez, Source | WEE1 homolog (S. pombe) | 11948 | -0.154 | -0.1281 | No |
| 319 | MME | MME Entrez, Source | membrane metallo-endopeptidase (neutral endopeptidase, enkephalinase) | 11986 | -0.156 | -0.1281 | No |
| 320 | AKAP11 | AKAP11 Entrez, Source | A kinase (PRKA) anchor protein 11 | 12060 | -0.162 | -0.1309 | No |
| 321 | AK3L1 | AK3L1 Entrez, Source | adenylate kinase 3-like 1 | 12108 | -0.165 | -0.1315 | No |
| 322 | PGK1 | PGK1 Entrez, Source | phosphoglycerate kinase 1 | 12194 | -0.171 | -0.1350 | No |
| 323 | FMR1 | FMR1 Entrez, Source | fragile X mental retardation 1 | 12198 | -0.171 | -0.1322 | No |
| 324 | VEGF | VEGF Entrez, Source | vascular endothelial growth factor | 12221 | -0.173 | -0.1308 | No |
| 325 | PDZK1 | PDZK1 Entrez, Source | PDZ domain containing 1 | 12224 | -0.173 | -0.1278 | No |
| 326 | NAT8 | NAT8 Entrez, Source | N-acetyltransferase 8 (camello like) | 12276 | -0.177 | -0.1286 | No |
| 327 | ST13 | ST13 Entrez, Source | suppression of tumorigenicity 13 (colon carcinoma) (Hsp70 interacting protein) | 12397 | -0.188 | -0.1345 | No |
| 328 | SDC1 | SDC1 Entrez, Source | syndecan 1 | 12438 | -0.192 | -0.1341 | No |
| 329 | EBNA1BP2 | EBNA1BP2 Entrez, Source | EBNA1 binding protein 2 | 12468 | -0.195 | -0.1328 | No |
| 330 | PRPSAP2 | PRPSAP2 Entrez, Source | phosphoribosyl pyrophosphate synthetase-associated protein 2 | 12563 | -0.206 | -0.1364 | No |
| 331 | PARD3 | PARD3 Entrez, Source | par-3 partitioning defective 3 homolog (C. elegans) | 12567 | -0.207 | -0.1329 | No |
| 332 | TOMM70A | TOMM70A Entrez, Source | translocase of outer mitochondrial membrane 70 homolog A (S. cerevisiae) | 12582 | -0.208 | -0.1303 | No |
| 333 | GGH | GGH Entrez, Source | gamma-glutamyl hydrolase (conjugase, folylpolygammaglutamyl hydrolase) | 12617 | -0.213 | -0.1291 | No |
| 334 | ATF2 | ATF2 Entrez, Source | activating transcription factor 2 | 12642 | -0.216 | -0.1270 | No |
| 335 | KIF3A | KIF3A Entrez, Source | kinesin family member 3A | 12663 | -0.219 | -0.1247 | No |
| 336 | SMAD5 | SMAD5 Entrez, Source | SMAD, mothers against DPP homolog 5 (Drosophila) | 12778 | -0.238 | -0.1292 | No |
| 337 | GBE1 | GBE1 Entrez, Source | glucan (1,4-alpha-), branching enzyme 1 (glycogen branching enzyme, Andersen disease, glycogen storage disease type IV) | 12796 | -0.240 | -0.1262 | No |
| 338 | FMO1 | FMO1 Entrez, Source | flavin containing monooxygenase 1 | 12848 | -0.253 | -0.1256 | No |
| 339 | VAT1 | VAT1 Entrez, Source | vesicle amine transport protein 1 homolog (T californica) | 12879 | -0.262 | -0.1232 | No |
| 340 | SLC34A1 | SLC34A1 Entrez, Source | solute carrier family 34 (sodium phosphate), member 1 | 12983 | -0.288 | -0.1260 | No |
| 341 | PDLIM5 | PDLIM5 Entrez, Source | PDZ and LIM domain 5 | 12986 | -0.289 | -0.1210 | No |
| 342 | SLC4A4 | SLC4A4 Entrez, Source | solute carrier family 4, sodium bicarbonate cotransporter, member 4 | 13008 | -0.294 | -0.1173 | No |
| 343 | SLC13A3 | SLC13A3 Entrez, Source | solute carrier family 13 (sodium-dependent dicarboxylate transporter), member 3 | 13078 | -0.318 | -0.1169 | No |
| 344 | HSPB1 | HSPB1 Entrez, Source | heat shock 27kDa protein 1 | 13125 | -0.341 | -0.1144 | No |
| 345 | UGDH | UGDH Entrez, Source | UDP-glucose dehydrogenase | 13138 | -0.352 | -0.1090 | No |
| 346 | CLIC4 | CLIC4 Entrez, Source | chloride intracellular channel 4 | 13159 | -0.370 | -0.1039 | No |
| 347 | ACSM3 | ACSM3 Entrez, Source | acyl-CoA synthetase medium-chain family member 3 | 13175 | -0.381 | -0.0983 | No |
| 348 | ENTPD5 | ENTPD5 Entrez, Source | ectonucleoside triphosphate diphosphohydrolase 5 | 13183 | -0.385 | -0.0919 | No |
| 349 | XPNPEP2 | XPNPEP2 Entrez, Source | X-prolyl aminopeptidase (aminopeptidase P) 2, membrane-bound | 13200 | -0.402 | -0.0859 | No |
| 350 | TNPO1 | TNPO1 Entrez, Source | transportin 1 | 13223 | -0.430 | -0.0799 | No |
| 351 | AKR7A3 | AKR7A3 Entrez, Source | aldo-keto reductase family 7, member A3 (aflatoxin aldehyde reductase) | 13236 | -0.447 | -0.0729 | No |
| 352 | NDRG1 | NDRG1 Entrez, Source | N-myc downstream regulated gene 1 | 13268 | -0.519 | -0.0660 | No |
| 353 | ABCC4 | ABCC4 Entrez, Source | ATP-binding cassette, sub-family C (CFTR/MRP), member 4 | 13272 | -0.523 | -0.0568 | No |
| 354 | SLC3A2 | SLC3A2 Entrez, Source | solute carrier family 3 (activators of dibasic and neutral amino acid transport), member 2 | 13274 | -0.526 | -0.0475 | No |
| 355 | SLC17A1 | SLC17A1 Entrez, Source | solute carrier family 17 (sodium phosphate), member 1 | 13290 | -0.561 | -0.0386 | No |
| 356 | GSTA2 | GSTA2 Entrez, Source | glutathione S-transferase A2 | 13300 | -0.608 | -0.0284 | No |
| 357 | SLC17A3 | SLC17A3 Entrez, Source | solute carrier family 17 (sodium phosphate), member 3 | 13320 | -0.761 | -0.0162 | No |
| 358 | RGN | RGN Entrez, Source | regucalcin (senescence marker protein-30) | 13333 | -0.998 | 0.0007 | No |