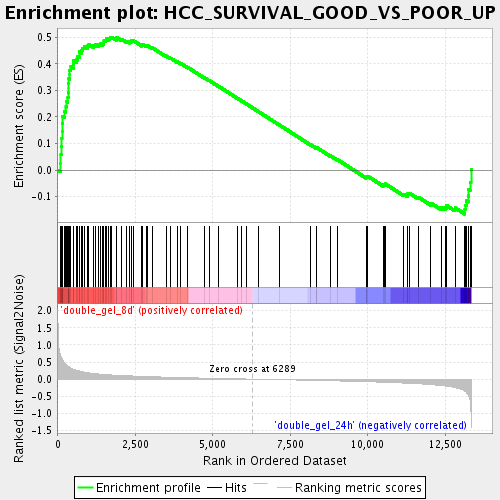
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | double_gel_and_single_gel_rma_expression_values_collapsed_to_symbols.class.cls #double_gel_8d_versus_double_gel_24h.class.cls #double_gel_8d_versus_double_gel_24h_repos |
| Phenotype | class.cls#double_gel_8d_versus_double_gel_24h_repos |
| Upregulated in class | double_gel_8d |
| GeneSet | HCC_SURVIVAL_GOOD_VS_POOR_UP |
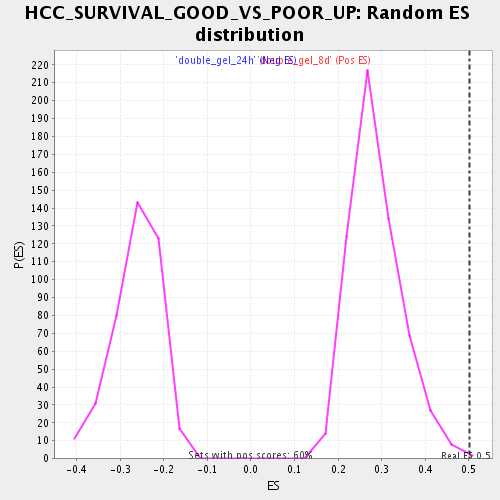
| Enrichment Score (ES) | 0.50038666 |
| Normalized Enrichment Score (NES) | 1.7444067 |
| Nominal p-value | 0.0016806723 |
| FDR q-value | 0.041447796 |
| FWER p-Value | 0.997 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | FMO3 | FMO3 Entrez, Source | flavin containing monooxygenase 3 | 94 | 0.673 | 0.0261 | Yes |
| 2 | ACOX2 | ACOX2 Entrez, Source | acyl-Coenzyme A oxidase 2, branched chain | 97 | 0.667 | 0.0589 | Yes |
| 3 | OTC | OTC Entrez, Source | ornithine carbamoyltransferase | 115 | 0.625 | 0.0885 | Yes |
| 4 | PCK1 | PCK1 Entrez, Source | phosphoenolpyruvate carboxykinase 1 (soluble) | 127 | 0.610 | 0.1177 | Yes |
| 5 | PAH | PAH Entrez, Source | phenylalanine hydroxylase | 133 | 0.597 | 0.1468 | Yes |
| 6 | HGD | HGD Entrez, Source | homogentisate 1,2-dioxygenase (homogentisate oxidase) | 138 | 0.591 | 0.1757 | Yes |
| 7 | CES2 | CES2 Entrez, Source | carboxylesterase 2 (intestine, liver) | 149 | 0.564 | 0.2028 | Yes |
| 8 | LPIN1 | LPIN1 Entrez, Source | lipin 1 | 216 | 0.473 | 0.2212 | Yes |
| 9 | ABHD6 | ABHD6 Entrez, Source | abhydrolase domain containing 6 | 260 | 0.429 | 0.2391 | Yes |
| 10 | DPYS | DPYS Entrez, Source | dihydropyrimidinase | 287 | 0.410 | 0.2574 | Yes |
| 11 | PC | PC Entrez, Source | pyruvate carboxylase | 322 | 0.380 | 0.2736 | Yes |
| 12 | SELENBP1 | SELENBP1 Entrez, Source | selenium binding protein 1 | 336 | 0.371 | 0.2909 | Yes |
| 13 | ECHS1 | ECHS1 Entrez, Source | enoyl Coenzyme A hydratase, short chain, 1, mitochondrial | 346 | 0.367 | 0.3084 | Yes |
| 14 | G6PC | G6PC Entrez, Source | glucose-6-phosphatase, catalytic subunit | 347 | 0.366 | 0.3265 | Yes |
| 15 | MUCDHL | MUCDHL Entrez, Source | mucin and cadherin-like | 353 | 0.363 | 0.3440 | Yes |
| 16 | EHHADH | EHHADH Entrez, Source | enoyl-Coenzyme A, hydratase/3-hydroxyacyl Coenzyme A dehydrogenase | 375 | 0.349 | 0.3596 | Yes |
| 17 | GLYAT | GLYAT Entrez, Source | glycine-N-acyltransferase | 379 | 0.348 | 0.3766 | Yes |
| 18 | CPT2 | CPT2 Entrez, Source | carnitine palmitoyltransferase II | 419 | 0.326 | 0.3897 | Yes |
| 19 | TTPA | TTPA Entrez, Source | tocopherol (alpha) transfer protein (ataxia (Friedreich-like) with vitamin E deficiency) | 495 | 0.288 | 0.3982 | Yes |
| 20 | SERPIND1 | SERPIND1 Entrez, Source | serpin peptidase inhibitor, clade D (heparin cofactor), member 1 | 504 | 0.284 | 0.4116 | Yes |
| 21 | BAAT | BAAT Entrez, Source | bile acid Coenzyme A: amino acid N-acyltransferase (glycine N-choloyltransferase) | 602 | 0.257 | 0.4170 | Yes |
| 22 | HADH2 | HADH2 Entrez, Source | hydroxyacyl-Coenzyme A dehydrogenase, type II | 638 | 0.247 | 0.4265 | Yes |
| 23 | CDO1 | CDO1 Entrez, Source | cysteine dioxygenase, type I | 689 | 0.237 | 0.4345 | Yes |
| 24 | GJB1 | GJB1 Entrez, Source | gap junction protein, beta 1, 32kDa (connexin 32, Charcot-Marie-Tooth neuropathy, X-linked) | 690 | 0.237 | 0.4462 | Yes |
| 25 | DCXR | DCXR Entrez, Source | dicarbonyl/L-xylulose reductase | 765 | 0.222 | 0.4516 | Yes |
| 26 | IVD | IVD Entrez, Source | isovaleryl Coenzyme A dehydrogenase | 800 | 0.213 | 0.4595 | Yes |
| 27 | MASP2 | MASP2 Entrez, Source | mannan-binding lectin serine peptidase 2 | 844 | 0.206 | 0.4664 | Yes |
| 28 | STARD10 | STARD10 Entrez, Source | START domain containing 10 | 953 | 0.188 | 0.4676 | Yes |
| 29 | MVK | MVK Entrez, Source | mevalonate kinase (mevalonic aciduria) | 998 | 0.183 | 0.4733 | Yes |
| 30 | PCCB | PCCB Entrez, Source | propionyl Coenzyme A carboxylase, beta polypeptide | 1152 | 0.165 | 0.4698 | Yes |
| 31 | IL6R | IL6R Entrez, Source | interleukin 6 receptor | 1216 | 0.160 | 0.4730 | Yes |
| 32 | SLC38A3 | SLC38A3 Entrez, Source | solute carrier family 38, member 3 | 1323 | 0.149 | 0.4723 | Yes |
| 33 | MSRA | MSRA Entrez, Source | methionine sulfoxide reductase A | 1379 | 0.144 | 0.4753 | Yes |
| 34 | KLK3 | KLK3 Entrez, Source | kallikrein 3, (prostate specific antigen) | 1448 | 0.139 | 0.4770 | Yes |
| 35 | CRAT | CRAT Entrez, Source | carnitine acetyltransferase | 1485 | 0.136 | 0.4810 | Yes |
| 36 | APOA5 | APOA5 Entrez, Source | apolipoprotein A-V | 1486 | 0.136 | 0.4877 | Yes |
| 37 | ACADS | ACADS Entrez, Source | acyl-Coenzyme A dehydrogenase, C-2 to C-3 short chain | 1544 | 0.132 | 0.4899 | Yes |
| 38 | NDUFS2 | NDUFS2 Entrez, Source | NADH dehydrogenase (ubiquinone) Fe-S protein 2, 49kDa (NADH-coenzyme Q reductase) | 1554 | 0.131 | 0.4957 | Yes |
| 39 | SLC2A2 | SLC2A2 Entrez, Source | solute carrier family 2 (facilitated glucose transporter), member 2 | 1645 | 0.126 | 0.4951 | Yes |
| 40 | INSIG1 | INSIG1 Entrez, Source | insulin induced gene 1 | 1681 | 0.124 | 0.4986 | Yes |
| 41 | CPB2 | CPB2 Entrez, Source | carboxypeptidase B2 (plasma, carboxypeptidase U) | 1738 | 0.120 | 0.5003 | Yes |
| 42 | APOC3 | APOC3 Entrez, Source | apolipoprotein C-III | 1882 | 0.113 | 0.4951 | Yes |
| 43 | AR | AR Entrez, Source | androgen receptor (dihydrotestosterone receptor; testicular feminization; spinal and bulbar muscular atrophy; Kennedy disease) | 1886 | 0.112 | 0.5004 | Yes |
| 44 | F12 | F12 Entrez, Source | coagulation factor XII (Hageman factor) | 2056 | 0.105 | 0.4928 | No |
| 45 | AMT | AMT Entrez, Source | aminomethyltransferase | 2221 | 0.098 | 0.4853 | No |
| 46 | PPAP2A | PPAP2A Entrez, Source | phosphatidic acid phosphatase type 2A | 2313 | 0.095 | 0.4831 | No |
| 47 | SERPINF1 | SERPINF1 Entrez, Source | serpin peptidase inhibitor, clade F (alpha-2 antiplasmin, pigment epithelium derived factor), member 1 | 2361 | 0.093 | 0.4842 | No |
| 48 | WNT11 | WNT11 Entrez, Source | wingless-type MMTV integration site family, member 11 | 2382 | 0.092 | 0.4872 | No |
| 49 | EGLN2 | EGLN2 Entrez, Source | egl nine homolog 2 (C. elegans) | 2444 | 0.090 | 0.4871 | No |
| 50 | ABCG5 | ABCG5 Entrez, Source | ATP-binding cassette, sub-family G (WHITE), member 5 (sterolin 1) | 2712 | 0.080 | 0.4708 | No |
| 51 | SLC6A12 | SLC6A12 Entrez, Source | solute carrier family 6 (neurotransmitter transporter, betaine/GABA), member 12 | 2727 | 0.079 | 0.4737 | No |
| 52 | ENPP5 | ENPP5 Entrez, Source | ectonucleotide pyrophosphatase/phosphodiesterase 5 (putative function) | 2843 | 0.076 | 0.4688 | No |
| 53 | MST1 | MST1 Entrez, Source | macrophage stimulating 1 (hepatocyte growth factor-like) | 2899 | 0.074 | 0.4683 | No |
| 54 | SERPING1 | SERPING1 Entrez, Source | serpin peptidase inhibitor, clade G (C1 inhibitor), member 1, (angioedema, hereditary) | 3039 | 0.070 | 0.4612 | No |
| 55 | PCYT2 | PCYT2 Entrez, Source | phosphate cytidylyltransferase 2, ethanolamine | 3493 | 0.058 | 0.4299 | No |
| 56 | HAGH | HAGH Entrez, Source | hydroxyacylglutathione hydrolase | 3639 | 0.054 | 0.4216 | No |
| 57 | MYRIP | MYRIP Entrez, Source | myosin VIIA and Rab interacting protein | 3846 | 0.049 | 0.4085 | No |
| 58 | SLC25A10 | SLC25A10 Entrez, Source | solute carrier family 25 (mitochondrial carrier; dicarboxylate transporter), member 10 | 3966 | 0.045 | 0.4017 | No |
| 59 | ACF | ACF Entrez, Source | - | 4189 | 0.040 | 0.3869 | No |
| 60 | ITPR2 | ITPR2 Entrez, Source | inositol 1,4,5-triphosphate receptor, type 2 | 4733 | 0.030 | 0.3474 | No |
| 61 | PRDX6 | PRDX6 Entrez, Source | peroxiredoxin 6 | 4883 | 0.026 | 0.3375 | No |
| 62 | SULT2A1 | SULT2A1 Entrez, Source | sulfotransferase family, cytosolic, 2A, dehydroepiandrosterone (DHEA)-preferring, member 1 | 5184 | 0.021 | 0.3158 | No |
| 63 | SERPINC1 | SERPINC1 Entrez, Source | serpin peptidase inhibitor, clade C (antithrombin), member 1 | 5806 | 0.009 | 0.2694 | No |
| 64 | AQP9 | AQP9 Entrez, Source | aquaporin 9 | 5937 | 0.006 | 0.2599 | No |
| 65 | ZP3 | ZP3 Entrez, Source | zona pellucida glycoprotein 3 (sperm receptor) | 6101 | 0.003 | 0.2478 | No |
| 66 | HPD | HPD Entrez, Source | 4-hydroxyphenylpyruvate dioxygenase | 6481 | -0.003 | 0.2193 | No |
| 67 | F10 | F10 Entrez, Source | coagulation factor X | 7144 | -0.015 | 0.1701 | No |
| 68 | ASGR1 | ASGR1 Entrez, Source | asialoglycoprotein receptor 1 | 8158 | -0.034 | 0.0953 | No |
| 69 | CRYL1 | CRYL1 Entrez, Source | crystallin, lambda 1 | 8339 | -0.038 | 0.0836 | No |
| 70 | CABIN1 | CABIN1 Entrez, Source | - | 8360 | -0.039 | 0.0840 | No |
| 71 | SEC14L2 | SEC14L2 Entrez, Source | SEC14-like 2 (S. cerevisiae) | 8781 | -0.047 | 0.0546 | No |
| 72 | CYP3A4 | CYP3A4 Entrez, Source | cytochrome P450, family 3, subfamily A, polypeptide 4 | 9007 | -0.052 | 0.0402 | No |
| 73 | NDRG2 | NDRG2 Entrez, Source | NDRG family member 2 | 9944 | -0.075 | -0.0268 | No |
| 74 | ABCC2 | ABCC2 Entrez, Source | ATP-binding cassette, sub-family C (CFTR/MRP), member 2 | 9965 | -0.075 | -0.0245 | No |
| 75 | ALAS1 | ALAS1 Entrez, Source | aminolevulinate, delta-, synthase 1 | 9995 | -0.076 | -0.0230 | No |
| 76 | ABCG8 | ABCG8 Entrez, Source | ATP-binding cassette, sub-family G (WHITE), member 8 (sterolin 2) | 10496 | -0.090 | -0.0563 | No |
| 77 | C2 | C2 Entrez, Source | complement component 2 | 10542 | -0.091 | -0.0551 | No |
| 78 | NF1 | NF1 Entrez, Source | neurofibromin 1 (neurofibromatosis, von Recklinghausen disease, Watson disease) | 10565 | -0.092 | -0.0522 | No |
| 79 | INSR | INSR Entrez, Source | insulin receptor | 11154 | -0.115 | -0.0910 | No |
| 80 | MRPL46 | MRPL46 Entrez, Source | mitochondrial ribosomal protein L46 | 11276 | -0.119 | -0.0942 | No |
| 81 | KHK | KHK Entrez, Source | ketohexokinase (fructokinase) | 11294 | -0.120 | -0.0896 | No |
| 82 | MPDZ | MPDZ Entrez, Source | multiple PDZ domain protein | 11344 | -0.122 | -0.0872 | No |
| 83 | APCS | APCS Entrez, Source | amyloid P component, serum | 11626 | -0.135 | -0.1018 | No |
| 84 | AMACR | AMACR Entrez, Source | alpha-methylacyl-CoA racemase | 12034 | -0.160 | -0.1246 | No |
| 85 | EPHX1 | EPHX1 Entrez, Source | epoxide hydrolase 1, microsomal (xenobiotic) | 12370 | -0.186 | -0.1407 | No |
| 86 | PKLR | PKLR Entrez, Source | pyruvate kinase, liver and RBC | 12501 | -0.200 | -0.1407 | No |
| 87 | CABC1 | CABC1 Entrez, Source | chaperone, ABC1 activity of bc1 complex homolog (S. pombe) | 12544 | -0.204 | -0.1338 | No |
| 88 | C4BPB | C4BPB Entrez, Source | complement component 4 binding protein, beta | 12817 | -0.245 | -0.1422 | No |
| 89 | CES1 | CES1 Entrez, Source | carboxylesterase 1 (monocyte/macrophage serine esterase 1) | 13135 | -0.348 | -0.1490 | No |
| 90 | SRD5A1 | SRD5A1 Entrez, Source | steroid-5-alpha-reductase, alpha polypeptide 1 (3-oxo-5 alpha-steroid delta 4-dehydrogenase alpha 1) | 13143 | -0.354 | -0.1320 | No |
| 91 | SGPL1 | SGPL1 Entrez, Source | sphingosine-1-phosphate lyase 1 | 13181 | -0.384 | -0.1159 | No |
| 92 | CES3 | CES3 Entrez, Source | carboxylesterase 3 (brain) | 13245 | -0.465 | -0.0976 | No |
| 93 | SLC27A5 | SLC27A5 Entrez, Source | solute carrier family 27 (fatty acid transporter), member 5 | 13261 | -0.509 | -0.0736 | No |
| 94 | AOX1 | AOX1 Entrez, Source | aldehyde oxidase 1 | 13304 | -0.615 | -0.0465 | No |
| 95 | RGN | RGN Entrez, Source | regucalcin (senescence marker protein-30) | 13333 | -0.998 | 0.0007 | No |