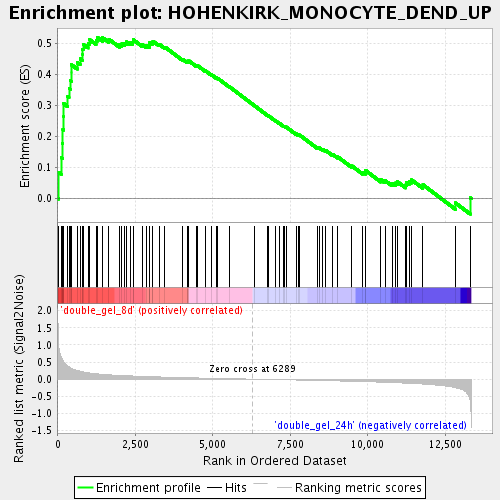
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | double_gel_and_single_gel_rma_expression_values_collapsed_to_symbols.class.cls #double_gel_8d_versus_double_gel_24h.class.cls #double_gel_8d_versus_double_gel_24h_repos |
| Phenotype | class.cls#double_gel_8d_versus_double_gel_24h_repos |
| Upregulated in class | double_gel_8d |
| GeneSet | HOHENKIRK_MONOCYTE_DEND_UP |
| Enrichment Score (ES) | 0.51852685 |
| Normalized Enrichment Score (NES) | 1.7786258 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.032630198 |
| FWER p-Value | 0.977 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | CCND2 | CCND2 Entrez, Source | cyclin D2 | 21 | 1.039 | 0.0849 | Yes |
| 2 | LPL | LPL Entrez, Source | lipoprotein lipase | 107 | 0.646 | 0.1323 | Yes |
| 3 | CBS | CBS Entrez, Source | cystathionine-beta-synthase | 148 | 0.566 | 0.1764 | Yes |
| 4 | TNFAIP6 | TNFAIP6 Entrez, Source | tumor necrosis factor, alpha-induced protein 6 | 159 | 0.550 | 0.2215 | Yes |
| 5 | ABCG1 | ABCG1 Entrez, Source | ATP-binding cassette, sub-family G (WHITE), member 1 | 179 | 0.519 | 0.2632 | Yes |
| 6 | F13A1 | F13A1 Entrez, Source | coagulation factor XIII, A1 polypeptide | 186 | 0.510 | 0.3053 | Yes |
| 7 | GDF15 | GDF15 Entrez, Source | growth differentiation factor 15 | 315 | 0.388 | 0.3279 | Yes |
| 8 | CYB5A | CYB5A Entrez, Source | cytochrome b5 type A (microsomal) | 359 | 0.359 | 0.3545 | Yes |
| 9 | PFKP | PFKP Entrez, Source | phosphofructokinase, platelet | 401 | 0.337 | 0.3795 | Yes |
| 10 | SCARB1 | SCARB1 Entrez, Source | scavenger receptor class B, member 1 | 431 | 0.321 | 0.4040 | Yes |
| 11 | HSD11B1 | HSD11B1 Entrez, Source | hydroxysteroid (11-beta) dehydrogenase 1 | 432 | 0.320 | 0.4307 | Yes |
| 12 | ACAA2 | ACAA2 Entrez, Source | acetyl-Coenzyme A acyltransferase 2 (mitochondrial 3-oxoacyl-Coenzyme A thiolase) | 624 | 0.251 | 0.4371 | Yes |
| 13 | MGLL | MGLL Entrez, Source | monoglyceride lipase | 718 | 0.232 | 0.4494 | Yes |
| 14 | C1QB | C1QB Entrez, Source | complement component 1, q subcomponent, B chain | 786 | 0.217 | 0.4624 | Yes |
| 15 | CITED2 | CITED2 Entrez, Source | Cbp/p300-interacting transactivator, with Glu/Asp-rich carboxy-terminal domain, 2 | 793 | 0.215 | 0.4799 | Yes |
| 16 | CD59 | CD59 Entrez, Source | CD59 molecule, complement regulatory protein | 818 | 0.210 | 0.4956 | Yes |
| 17 | ATP1B1 | ATP1B1 Entrez, Source | ATPase, Na+/K+ transporting, beta 1 polypeptide | 973 | 0.186 | 0.4995 | Yes |
| 18 | BIRC3 | BIRC3 Entrez, Source | baculoviral IAP repeat-containing 3 | 1025 | 0.179 | 0.5105 | Yes |
| 19 | RNASE6 | RNASE6 Entrez, Source | ribonuclease, RNase A family, k6 | 1232 | 0.158 | 0.5082 | Yes |
| 20 | DUSP1 | DUSP1 Entrez, Source | dual specificity phosphatase 1 | 1266 | 0.154 | 0.5185 | Yes |
| 21 | TGFA | TGFA Entrez, Source | transforming growth factor, alpha | 1441 | 0.140 | 0.5170 | No |
| 22 | CSF1 | CSF1 Entrez, Source | colony stimulating factor 1 (macrophage) | 1644 | 0.126 | 0.5123 | No |
| 23 | AES | AES Entrez, Source | amino-terminal enhancer of split | 1989 | 0.108 | 0.4954 | No |
| 24 | CCL17 | CCL17 Entrez, Source | chemokine (C-C motif) ligand 17 | 2066 | 0.105 | 0.4984 | No |
| 25 | MAOA | MAOA Entrez, Source | monoamine oxidase A | 2159 | 0.101 | 0.4999 | No |
| 26 | ALOX15 | ALOX15 Entrez, Source | arachidonate 15-lipoxygenase | 2217 | 0.099 | 0.5038 | No |
| 27 | SPP1 | SPP1 Entrez, Source | secreted phosphoprotein 1 (osteopontin, bone sialoprotein I, early T-lymphocyte activation 1) | 2345 | 0.094 | 0.5020 | No |
| 28 | FABP3 | FABP3 Entrez, Source | fatty acid binding protein 3, muscle and heart (mammary-derived growth inhibitor) | 2423 | 0.091 | 0.5037 | No |
| 29 | PPIC | PPIC Entrez, Source | peptidylprolyl isomerase C (cyclophilin C) | 2432 | 0.090 | 0.5106 | No |
| 30 | ECM1 | ECM1 Entrez, Source | extracellular matrix protein 1 | 2718 | 0.080 | 0.4958 | No |
| 31 | CDH1 | CDH1 Entrez, Source | cadherin 1, type 1, E-cadherin (epithelial) | 2848 | 0.076 | 0.4923 | No |
| 32 | ENPP2 | ENPP2 Entrez, Source | ectonucleotide pyrophosphatase/phosphodiesterase 2 (autotaxin) | 2952 | 0.072 | 0.4906 | No |
| 33 | HMGN3 | HMGN3 Entrez, Source | high mobility group nucleosomal binding domain 3 | 2956 | 0.072 | 0.4964 | No |
| 34 | ARF4 | ARF4 Entrez, Source | ADP-ribosylation factor 4 | 2963 | 0.072 | 0.5019 | No |
| 35 | GAS6 | GAS6 Entrez, Source | growth arrest-specific 6 | 3043 | 0.070 | 0.5018 | No |
| 36 | SPINT2 | SPINT2 Entrez, Source | serine peptidase inhibitor, Kunitz type, 2 | 3064 | 0.069 | 0.5061 | No |
| 37 | CCNA1 | CCNA1 Entrez, Source | cyclin A1 | 3266 | 0.064 | 0.4962 | No |
| 38 | ST6GAL1 | ST6GAL1 Entrez, Source | ST6 beta-galactosamide alpha-2,6-sialyltranferase 1 | 3452 | 0.059 | 0.4872 | No |
| 39 | APOC1 | APOC1 Entrez, Source | apolipoprotein C-I | 4022 | 0.044 | 0.4479 | No |
| 40 | PFKM | PFKM Entrez, Source | phosphofructokinase, muscle | 4179 | 0.040 | 0.4395 | No |
| 41 | SULT1C1 | SULT1C1 Entrez, Source | sulfotransferase family, cytosolic, 1C, member 1 | 4188 | 0.040 | 0.4422 | No |
| 42 | NR1H3 | NR1H3 Entrez, Source | nuclear receptor subfamily 1, group H, member 3 | 4228 | 0.039 | 0.4425 | No |
| 43 | CTSC | CTSC Entrez, Source | cathepsin C | 4468 | 0.035 | 0.4274 | No |
| 44 | ACOT7 | ACOT7 Entrez, Source | acyl-CoA thioesterase 7 | 4493 | 0.034 | 0.4284 | No |
| 45 | NR4A3 | NR4A3 Entrez, Source | nuclear receptor subfamily 4, group A, member 3 | 4753 | 0.029 | 0.4113 | No |
| 46 | A2M | A2M Entrez, Source | alpha-2-macroglobulin | 4951 | 0.025 | 0.3985 | No |
| 47 | RAP1GAP | RAP1GAP Entrez, Source | RAP1 GTPase activating protein | 5106 | 0.022 | 0.3887 | No |
| 48 | ACOT2 | ACOT2 Entrez, Source | acyl-CoA thioesterase 2 | 5165 | 0.021 | 0.3861 | No |
| 49 | LGALS3BP | LGALS3BP Entrez, Source | lectin, galactoside-binding, soluble, 3 binding protein | 5525 | 0.014 | 0.3602 | No |
| 50 | DPP4 | DPP4 Entrez, Source | dipeptidyl-peptidase 4 (CD26, adenosine deaminase complexing protein 2) | 6342 | -0.001 | 0.2987 | No |
| 51 | PCBD1 | PCBD1 Entrez, Source | pterin-4 alpha-carbinolamine dehydratase/dimerization cofactor of hepatocyte nuclear factor 1 alpha (TCF1) | 6764 | -0.008 | 0.2677 | No |
| 52 | PDLIM4 | PDLIM4 Entrez, Source | PDZ and LIM domain 4 | 6797 | -0.009 | 0.2660 | No |
| 53 | SNRPN | SNRPN Entrez, Source | small nuclear ribonucleoprotein polypeptide N | 7006 | -0.012 | 0.2513 | No |
| 54 | CCL22 | CCL22 Entrez, Source | chemokine (C-C motif) ligand 22 | 7142 | -0.015 | 0.2424 | No |
| 55 | ITGB5 | ITGB5 Entrez, Source | integrin, beta 5 | 7282 | -0.018 | 0.2333 | No |
| 56 | ITPKB | ITPKB Entrez, Source | inositol 1,4,5-trisphosphate 3-kinase B | 7323 | -0.018 | 0.2319 | No |
| 57 | CRABP2 | CRABP2 Entrez, Source | cellular retinoic acid binding protein 2 | 7366 | -0.019 | 0.2303 | No |
| 58 | CD86 | CD86 Entrez, Source | CD86 molecule | 7686 | -0.025 | 0.2083 | No |
| 59 | INHBA | INHBA Entrez, Source | inhibin, beta A (activin A, activin AB alpha polypeptide) | 7768 | -0.027 | 0.2044 | No |
| 60 | SLC11A2 | SLC11A2 Entrez, Source | solute carrier family 11 (proton-coupled divalent metal ion transporters), member 2 | 7794 | -0.027 | 0.2048 | No |
| 61 | MMP12 | MMP12 Entrez, Source | matrix metallopeptidase 12 (macrophage elastase) | 8384 | -0.039 | 0.1636 | No |
| 62 | PTPRO | PTPRO Entrez, Source | protein tyrosine phosphatase, receptor type, O | 8433 | -0.040 | 0.1633 | No |
| 63 | DUSP5 | DUSP5 Entrez, Source | dual specificity phosphatase 5 | 8551 | -0.042 | 0.1581 | No |
| 64 | TOMM40 | TOMM40 Entrez, Source | translocase of outer mitochondrial membrane 40 homolog (yeast) | 8638 | -0.044 | 0.1553 | No |
| 65 | CAT | CAT Entrez, Source | catalase | 8873 | -0.049 | 0.1417 | No |
| 66 | DNASE1L3 | DNASE1L3 Entrez, Source | deoxyribonuclease I-like 3 | 9028 | -0.052 | 0.1345 | No |
| 67 | KCNAB2 | KCNAB2 Entrez, Source | potassium voltage-gated channel, shaker-related subfamily, beta member 2 | 9487 | -0.063 | 0.1051 | No |
| 68 | PEBP1 | PEBP1 Entrez, Source | phosphatidylethanolamine binding protein 1 | 9842 | -0.072 | 0.0844 | No |
| 69 | SLA | SLA Entrez, Source | Src-like-adaptor | 9920 | -0.074 | 0.0848 | No |
| 70 | RARRES1 | RARRES1 Entrez, Source | retinoic acid receptor responder (tazarotene induced) 1 | 9927 | -0.075 | 0.0906 | No |
| 71 | QSCN6 | QSCN6 Entrez, Source | quiescin Q6 | 10424 | -0.088 | 0.0605 | No |
| 72 | INDO | INDO Entrez, Source | indoleamine-pyrrole 2,3 dioxygenase | 10558 | -0.092 | 0.0581 | No |
| 73 | GATM | GATM Entrez, Source | glycine amidinotransferase (L-arginine:glycine amidinotransferase) | 10782 | -0.100 | 0.0497 | No |
| 74 | CCNH | CCNH Entrez, Source | cyclin H | 10890 | -0.105 | 0.0503 | No |
| 75 | TGM2 | TGM2 Entrez, Source | transglutaminase 2 (C polypeptide, protein-glutamine-gamma-glutamyltransferase) | 10955 | -0.107 | 0.0544 | No |
| 76 | FABP5 | FABP5 Entrez, Source | fatty acid binding protein 5 (psoriasis-associated) | 11225 | -0.117 | 0.0438 | No |
| 77 | TFRC | TFRC Entrez, Source | transferrin receptor (p90, CD71) | 11249 | -0.118 | 0.0519 | No |
| 78 | FCGR2B | FCGR2B Entrez, Source | Fc fragment of IgG, low affinity IIb, receptor (CD32) | 11337 | -0.122 | 0.0555 | No |
| 79 | FABP4 | FABP4 Entrez, Source | fatty acid binding protein 4, adipocyte | 11412 | -0.125 | 0.0603 | No |
| 80 | OAT | OAT Entrez, Source | ornithine aminotransferase (gyrate atrophy) | 11781 | -0.143 | 0.0445 | No |
| 81 | CEBPA | CEBPA Entrez, Source | CCAAT/enhancer binding protein (C/EBP), alpha | 12827 | -0.248 | -0.0136 | No |
| 82 | CD74 | CD74 Entrez, Source | CD74 molecule, major histocompatibility complex, class II invariant chain | 13307 | -0.629 | 0.0026 | No |