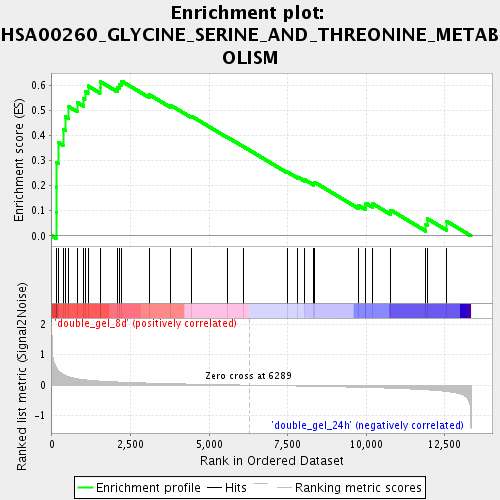
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | double_gel_and_single_gel_rma_expression_values_collapsed_to_symbols.class.cls #double_gel_8d_versus_double_gel_24h.class.cls #double_gel_8d_versus_double_gel_24h_repos |
| Phenotype | class.cls#double_gel_8d_versus_double_gel_24h_repos |
| Upregulated in class | double_gel_8d |
| GeneSet | HSA00260_GLYCINE_SERINE_AND_THREONINE_METABOLISM |
| Enrichment Score (ES) | 0.61707145 |
| Normalized Enrichment Score (NES) | 1.7915878 |
| Nominal p-value | 0.0017793594 |
| FDR q-value | 0.030322276 |
| FWER p-Value | 0.954 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | GNMT | GNMT Entrez, Source | glycine N-methyltransferase | 134 | 0.597 | 0.0942 | Yes |
| 2 | PEMT | PEMT Entrez, Source | phosphatidylethanolamine N-methyltransferase | 142 | 0.579 | 0.1949 | Yes |
| 3 | CBS | CBS Entrez, Source | cystathionine-beta-synthase | 148 | 0.566 | 0.2935 | Yes |
| 4 | AGXT | AGXT Entrez, Source | alanine-glyoxylate aminotransferase (oxalosis I; hyperoxaluria I; glycolicaciduria; serine-pyruvate aminotransferase) | 212 | 0.475 | 0.3717 | Yes |
| 5 | DMGDH | DMGDH Entrez, Source | dimethylglycine dehydrogenase | 357 | 0.361 | 0.4240 | Yes |
| 6 | AGXT2 | AGXT2 Entrez, Source | alanine-glyoxylate aminotransferase 2 | 428 | 0.322 | 0.4750 | Yes |
| 7 | CHDH | CHDH Entrez, Source | choline dehydrogenase | 528 | 0.275 | 0.5157 | Yes |
| 8 | PHGDH | PHGDH Entrez, Source | phosphoglycerate dehydrogenase | 810 | 0.211 | 0.5315 | Yes |
| 9 | SDS | SDS Entrez, Source | serine dehydratase | 1007 | 0.182 | 0.5486 | Yes |
| 10 | BHMT | BHMT Entrez, Source | betaine-homocysteine methyltransferase | 1054 | 0.177 | 0.5760 | Yes |
| 11 | SARDH | SARDH Entrez, Source | sarcosine dehydrogenase | 1150 | 0.165 | 0.5977 | Yes |
| 12 | MAOB | MAOB Entrez, Source | monoamine oxidase B | 1531 | 0.133 | 0.5924 | Yes |
| 13 | GAMT | GAMT Entrez, Source | guanidinoacetate N-methyltransferase | 1535 | 0.132 | 0.6153 | Yes |
| 14 | CTH | CTH Entrez, Source | cystathionase (cystathionine gamma-lyase) | 2088 | 0.104 | 0.5920 | Yes |
| 15 | MAOA | MAOA Entrez, Source | monoamine oxidase A | 2159 | 0.101 | 0.6044 | Yes |
| 16 | AMT | AMT Entrez, Source | aminomethyltransferase | 2221 | 0.098 | 0.6171 | Yes |
| 17 | ALAS2 | ALAS2 Entrez, Source | aminolevulinate, delta-, synthase 2 (sideroblastic/hypochromic anemia) | 3103 | 0.068 | 0.5628 | No |
| 18 | ABP1 | ABP1 Entrez, Source | amiloride binding protein 1 (amine oxidase (copper-containing)) | 3774 | 0.051 | 0.5213 | No |
| 19 | PSAT1 | PSAT1 Entrez, Source | phosphoserine aminotransferase 1 | 4440 | 0.035 | 0.4775 | No |
| 20 | AOC3 | AOC3 Entrez, Source | amine oxidase, copper containing 3 (vascular adhesion protein 1) | 5594 | 0.013 | 0.3931 | No |
| 21 | DLD | DLD Entrez, Source | dihydrolipoamide dehydrogenase | 6088 | 0.003 | 0.3566 | No |
| 22 | RDH11 | RDH11 Entrez, Source | retinol dehydrogenase 11 (all-trans/9-cis/11-cis) | 7495 | -0.022 | 0.2548 | No |
| 23 | SHMT1 | SHMT1 Entrez, Source | serine hydroxymethyltransferase 1 (soluble) | 7827 | -0.028 | 0.2348 | No |
| 24 | CHKB | CHKB Entrez, Source | choline kinase beta | 8048 | -0.032 | 0.2239 | No |
| 25 | HSD3B7 | HSD3B7 Entrez, Source | hydroxy-delta-5-steroid dehydrogenase, 3 beta- and steroid delta-isomerase 7 | 8340 | -0.038 | 0.2087 | No |
| 26 | AKR1B10 | AKR1B10 Entrez, Source | aldo-keto reductase family 1, member B10 (aldose reductase) | 8361 | -0.039 | 0.2139 | No |
| 27 | PSPH | PSPH Entrez, Source | phosphoserine phosphatase | 9762 | -0.069 | 0.1209 | No |
| 28 | GARS | GARS Entrez, Source | glycyl-tRNA synthetase | 9975 | -0.076 | 0.1182 | No |
| 29 | ALAS1 | ALAS1 Entrez, Source | aminolevulinate, delta-, synthase 1 | 9995 | -0.076 | 0.1301 | No |
| 30 | TARS | TARS Entrez, Source | threonyl-tRNA synthetase | 10218 | -0.082 | 0.1278 | No |
| 31 | GATM | GATM Entrez, Source | glycine amidinotransferase (L-arginine:glycine amidinotransferase) | 10782 | -0.100 | 0.1030 | No |
| 32 | SHMT2 | SHMT2 Entrez, Source | serine hydroxymethyltransferase 2 (mitochondrial) | 11904 | -0.150 | 0.0451 | No |
| 33 | CHKA | CHKA Entrez, Source | choline kinase alpha | 11951 | -0.154 | 0.0684 | No |
| 34 | GCAT | GCAT Entrez, Source | glycine C-acetyltransferase (2-amino-3-ketobutyrate coenzyme A ligase) | 12561 | -0.206 | 0.0587 | No |