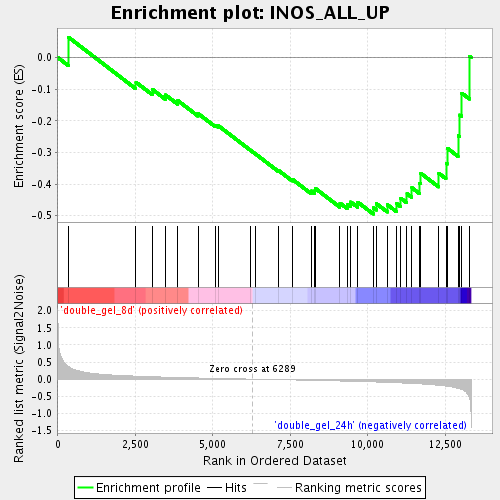
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | double_gel_and_single_gel_rma_expression_values_collapsed_to_symbols.class.cls #double_gel_8d_versus_double_gel_24h.class.cls #double_gel_8d_versus_double_gel_24h_repos |
| Phenotype | class.cls#double_gel_8d_versus_double_gel_24h_repos |
| Upregulated in class | double_gel_24h |
| GeneSet | INOS_ALL_UP |
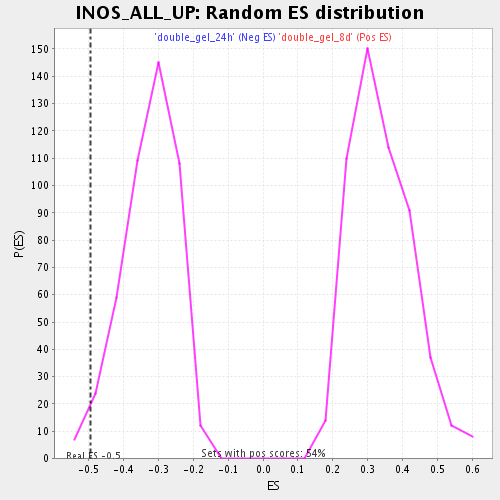
| Enrichment Score (ES) | -0.4949148 |
| Normalized Enrichment Score (NES) | -1.5237547 |
| Nominal p-value | 0.028017242 |
| FDR q-value | 0.22338344 |
| FWER p-Value | 1.0 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | TMSB4X | TMSB4X Entrez, Source | thymosin, beta 4, X-linked | 342 | 0.369 | 0.0644 | No |
| 2 | IER2 | IER2 Entrez, Source | immediate early response 2 | 2518 | 0.087 | -0.0777 | No |
| 3 | CCNE1 | CCNE1 Entrez, Source | cyclin E1 | 3049 | 0.070 | -0.1005 | No |
| 4 | IFRD1 | IFRD1 Entrez, Source | interferon-related developmental regulator 1 | 3472 | 0.058 | -0.1179 | No |
| 5 | ACLY | ACLY Entrez, Source | ATP citrate lyase | 3867 | 0.048 | -0.1357 | No |
| 6 | CSRP1 | CSRP1 Entrez, Source | cysteine and glycine-rich protein 1 | 4522 | 0.033 | -0.1767 | No |
| 7 | PICK1 | PICK1 Entrez, Source | protein interacting with PRKCA 1 | 5088 | 0.022 | -0.2137 | No |
| 8 | CSN3 | CSN3 Entrez, Source | casein kappa | 5180 | 0.021 | -0.2155 | No |
| 9 | SMAD2 | SMAD2 Entrez, Source | SMAD, mothers against DPP homolog 2 (Drosophila) | 6213 | 0.001 | -0.2928 | No |
| 10 | H2AFZ | H2AFZ Entrez, Source | H2A histone family, member Z | 6380 | -0.001 | -0.3049 | No |
| 11 | CDK7 | CDK7 Entrez, Source | cyclin-dependent kinase 7 (MO15 homolog, Xenopus laevis, cdk-activating kinase) | 7106 | -0.014 | -0.3560 | No |
| 12 | YWHAH | YWHAH Entrez, Source | tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, eta polypeptide | 7564 | -0.023 | -0.3847 | No |
| 13 | CDK2 | CDK2 Entrez, Source | cyclin-dependent kinase 2 | 8169 | -0.035 | -0.4217 | No |
| 14 | PCSK7 | PCSK7 Entrez, Source | proprotein convertase subtilisin/kexin type 7 | 8290 | -0.037 | -0.4216 | No |
| 15 | NEDD8 | NEDD8 Entrez, Source | neural precursor cell expressed, developmentally down-regulated 8 | 8297 | -0.037 | -0.4129 | No |
| 16 | RAC1 | RAC1 Entrez, Source | ras-related C3 botulinum toxin substrate 1 (rho family, small GTP binding protein Rac1) | 9101 | -0.054 | -0.4600 | No |
| 17 | RPL17 | RPL17 Entrez, Source | ribosomal protein L17 | 9343 | -0.059 | -0.4637 | No |
| 18 | HSPA8 | HSPA8 Entrez, Source | heat shock 70kDa protein 8 | 9449 | -0.062 | -0.4565 | No |
| 19 | TYMS | TYMS Entrez, Source | thymidylate synthetase | 9675 | -0.067 | -0.4570 | No |
| 20 | RBBP7 | RBBP7 Entrez, Source | retinoblastoma binding protein 7 | 10181 | -0.081 | -0.4751 | Yes |
| 21 | PABPC4 | PABPC4 Entrez, Source | poly(A) binding protein, cytoplasmic 4 (inducible form) | 10277 | -0.084 | -0.4618 | Yes |
| 22 | RPS12 | RPS12 Entrez, Source | ribosomal protein S12 | 10633 | -0.095 | -0.4653 | Yes |
| 23 | SRM | SRM Entrez, Source | spermidine synthase | 10934 | -0.106 | -0.4619 | Yes |
| 24 | ADM | ADM Entrez, Source | adrenomedullin | 11063 | -0.110 | -0.4445 | Yes |
| 25 | GTF2F1 | GTF2F1 Entrez, Source | general transcription factor IIF, polypeptide 1, 74kDa | 11265 | -0.119 | -0.4305 | Yes |
| 26 | RAN | RAN Entrez, Source | RAN, member RAS oncogene family | 11423 | -0.126 | -0.4116 | Yes |
| 27 | ATP6V1C1 | ATP6V1C1 Entrez, Source | ATPase, H+ transporting, lysosomal 42kDa, V1 subunit C1 | 11674 | -0.137 | -0.3969 | Yes |
| 28 | IL1RN | IL1RN Entrez, Source | interleukin 1 receptor antagonist | 11709 | -0.139 | -0.3656 | Yes |
| 29 | MIF | MIF Entrez, Source | macrophage migration inhibitory factor (glycosylation-inhibiting factor) | 12291 | -0.179 | -0.3656 | Yes |
| 30 | SMS | SMS Entrez, Source | spermine synthase | 12527 | -0.202 | -0.3339 | Yes |
| 31 | PDCD2 | PDCD2 Entrez, Source | programmed cell death 2 | 12564 | -0.206 | -0.2862 | Yes |
| 32 | LIG1 | LIG1 Entrez, Source | ligase I, DNA, ATP-dependent | 12917 | -0.270 | -0.2466 | Yes |
| 33 | FEN1 | FEN1 Entrez, Source | flap structure-specific endonuclease 1 | 12971 | -0.283 | -0.1814 | Yes |
| 34 | HSPH1 | HSPH1 Entrez, Source | heat shock 105kDa/110kDa protein 1 | 13031 | -0.302 | -0.1120 | Yes |
| 35 | MCM6 | MCM6 Entrez, Source | MCM6 minichromosome maintenance deficient 6 (MIS5 homolog, S. pombe) (S. cerevisiae) | 13288 | -0.553 | 0.0041 | Yes |