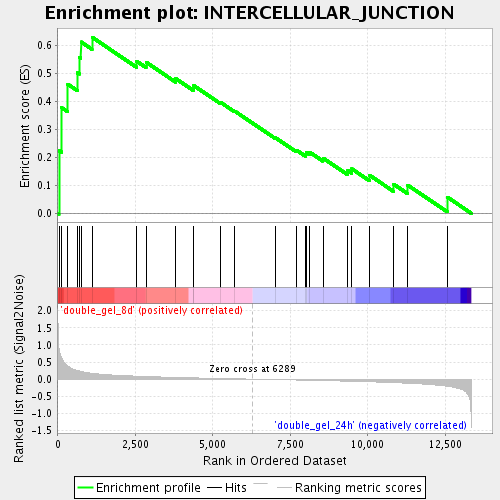
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | double_gel_and_single_gel_rma_expression_values_collapsed_to_symbols.class.cls #double_gel_8d_versus_double_gel_24h.class.cls #double_gel_8d_versus_double_gel_24h_repos |
| Phenotype | class.cls#double_gel_8d_versus_double_gel_24h_repos |
| Upregulated in class | double_gel_8d |
| GeneSet | INTERCELLULAR_JUNCTION |
| Enrichment Score (ES) | 0.63043517 |
| Normalized Enrichment Score (NES) | 1.7432244 |
| Nominal p-value | 0.0074766357 |
| FDR q-value | 0.04136264 |
| FWER p-Value | 0.998 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | CLDN7 | CLDN7 Entrez, Source | claudin 7 | 36 | 0.888 | 0.2247 | Yes |
| 2 | CLDN1 | CLDN1 Entrez, Source | claudin 1 | 118 | 0.623 | 0.3783 | Yes |
| 3 | GJA1 | GJA1 Entrez, Source | gap junction protein, alpha 1, 43kDa (connexin 43) | 316 | 0.387 | 0.4626 | Yes |
| 4 | GJB2 | GJB2 Entrez, Source | gap junction protein, beta 2, 26kDa (connexin 26) | 628 | 0.250 | 0.5032 | Yes |
| 5 | GJB1 | GJB1 Entrez, Source | gap junction protein, beta 1, 32kDa (connexin 32, Charcot-Marie-Tooth neuropathy, X-linked) | 690 | 0.237 | 0.5593 | Yes |
| 6 | CLDN3 | CLDN3 Entrez, Source | claudin 3 | 745 | 0.228 | 0.6137 | Yes |
| 7 | WNK4 | WNK4 Entrez, Source | WNK lysine deficient protein kinase 4 | 1104 | 0.170 | 0.6304 | Yes |
| 8 | CNKSR1 | CNKSR1 Entrez, Source | connector enhancer of kinase suppressor of Ras 1 | 2540 | 0.086 | 0.5447 | No |
| 9 | CDH1 | CDH1 Entrez, Source | cadherin 1, type 1, E-cadherin (epithelial) | 2848 | 0.076 | 0.5410 | No |
| 10 | CLDN19 | CLDN19 Entrez, Source | claudin 19 | 3802 | 0.050 | 0.4823 | No |
| 11 | PARD6A | PARD6A Entrez, Source | par-6 partitioning defective 6 homolog alpha (C.elegans) | 4374 | 0.036 | 0.4487 | No |
| 12 | CLDN8 | CLDN8 Entrez, Source | claudin 8 | 4382 | 0.036 | 0.4575 | No |
| 13 | CLDN14 | CLDN14 Entrez, Source | claudin 14 | 5251 | 0.020 | 0.3973 | No |
| 14 | DLG1 | DLG1 Entrez, Source | discs, large homolog 1 (Drosophila) | 5702 | 0.011 | 0.3663 | No |
| 15 | RAB13 | RAB13 Entrez, Source | RAB13, member RAS oncogene family | 7013 | -0.013 | 0.2712 | No |
| 16 | CLDN11 | CLDN11 Entrez, Source | claudin 11 (oligodendrocyte transmembrane protein) | 7693 | -0.025 | 0.2266 | No |
| 17 | CLDN4 | CLDN4 Entrez, Source | claudin 4 | 8005 | -0.031 | 0.2113 | No |
| 18 | CDC42BPA | CDC42BPA Entrez, Source | CDC42 binding protein kinase alpha (DMPK-like) | 8008 | -0.031 | 0.2191 | No |
| 19 | CLDN5 | CLDN5 Entrez, Source | claudin 5 (transmembrane protein deleted in velocardiofacial syndrome) | 8120 | -0.034 | 0.2195 | No |
| 20 | VAPA | VAPA Entrez, Source | VAMP (vesicle-associated membrane protein)-associated protein A, 33kDa | 8555 | -0.043 | 0.1978 | No |
| 21 | CDC42BPB | CDC42BPB Entrez, Source | CDC42 binding protein kinase beta (DMPK-like) | 9353 | -0.059 | 0.1531 | No |
| 22 | PDZD2 | PDZD2 Entrez, Source | PDZ domain containing 2 | 9485 | -0.063 | 0.1593 | No |
| 23 | CLDN16 | CLDN16 Entrez, Source | claudin 16 | 10062 | -0.078 | 0.1361 | No |
| 24 | ZYX | ZYX Entrez, Source | zyxin | 10834 | -0.102 | 0.1044 | No |
| 25 | MLLT4 | MLLT4 Entrez, Source | myeloid/lymphoid or mixed-lineage leukemia (trithorax homolog, Drosophila); translocated to, 4 | 11289 | -0.120 | 0.1011 | No |
| 26 | PARD3 | PARD3 Entrez, Source | par-3 partitioning defective 3 homolog (C. elegans) | 12567 | -0.207 | 0.0582 | No |