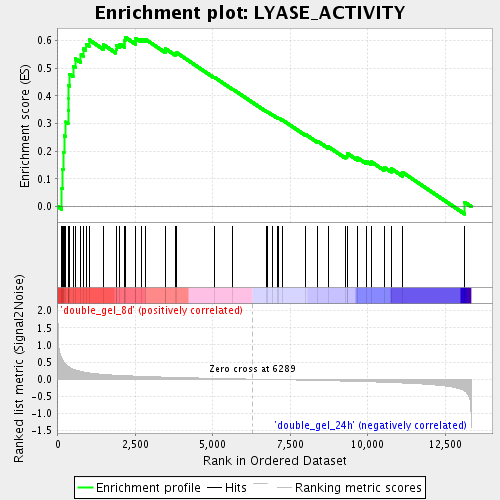
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | double_gel_and_single_gel_rma_expression_values_collapsed_to_symbols.class.cls #double_gel_8d_versus_double_gel_24h.class.cls #double_gel_8d_versus_double_gel_24h_repos |
| Phenotype | class.cls#double_gel_8d_versus_double_gel_24h_repos |
| Upregulated in class | double_gel_8d |
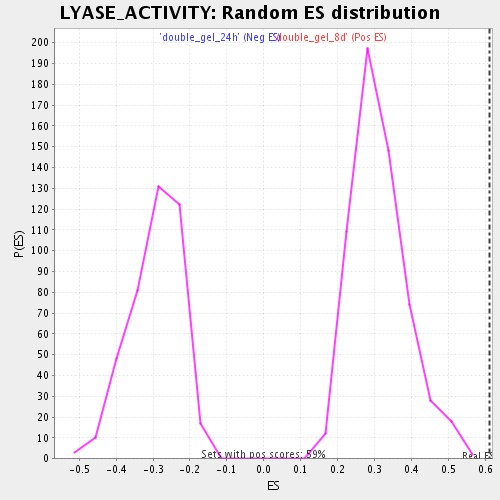
| GeneSet | LYASE_ACTIVITY |
| Enrichment Score (ES) | 0.6116802 |
| Normalized Enrichment Score (NES) | 1.946644 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.008298882 |
| FWER p-Value | 0.313 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | PCK1 | PCK1 Entrez, Source | phosphoenolpyruvate carboxykinase 1 (soluble) | 127 | 0.610 | 0.0665 | Yes |
| 2 | CBS | CBS Entrez, Source | cystathionine-beta-synthase | 148 | 0.566 | 0.1356 | Yes |
| 3 | GUCY2C | GUCY2C Entrez, Source | guanylate cyclase 2C (heat stable enterotoxin receptor) | 184 | 0.511 | 0.1967 | Yes |
| 4 | CA5A | CA5A Entrez, Source | carbonic anhydrase VA, mitochondrial | 199 | 0.487 | 0.2564 | Yes |
| 5 | BCKDHA | BCKDHA Entrez, Source | branched chain keto acid dehydrogenase E1, alpha polypeptide | 258 | 0.431 | 0.3058 | Yes |
| 6 | ACMSD | ACMSD Entrez, Source | aminocarboxymuconate semialdehyde decarboxylase | 328 | 0.374 | 0.3472 | Yes |
| 7 | ECHS1 | ECHS1 Entrez, Source | enoyl Coenzyme A hydratase, short chain, 1, mitochondrial | 346 | 0.367 | 0.3917 | Yes |
| 8 | ECH1 | ECH1 Entrez, Source | enoyl Coenzyme A hydratase 1, peroxisomal | 350 | 0.365 | 0.4370 | Yes |
| 9 | EHHADH | EHHADH Entrez, Source | enoyl-Coenzyme A, hydratase/3-hydroxyacyl Coenzyme A dehydrogenase | 375 | 0.349 | 0.4787 | Yes |
| 10 | HACL1 | HACL1 Entrez, Source | 2-hydroxyacyl-CoA lyase 1 | 490 | 0.289 | 0.5062 | Yes |
| 11 | GGCX | GGCX Entrez, Source | gamma-glutamyl carboxylase | 563 | 0.266 | 0.5339 | Yes |
| 12 | GUCY1B3 | GUCY1B3 Entrez, Source | guanylate cyclase 1, soluble, beta 3 | 744 | 0.228 | 0.5488 | Yes |
| 13 | MLYCD | MLYCD Entrez, Source | malonyl-CoA decarboxylase | 817 | 0.210 | 0.5696 | Yes |
| 14 | BCKDHB | BCKDHB Entrez, Source | branched chain keto acid dehydrogenase E1, beta polypeptide (maple syrup urine disease) | 906 | 0.195 | 0.5873 | Yes |
| 15 | SDS | SDS Entrez, Source | serine dehydratase | 1007 | 0.182 | 0.6025 | Yes |
| 16 | PTS | PTS Entrez, Source | 6-pyruvoyltetrahydropterin synthase | 1478 | 0.137 | 0.5842 | Yes |
| 17 | CRYM | CRYM Entrez, Source | crystallin, mu | 1874 | 0.113 | 0.5686 | Yes |
| 18 | ENO2 | ENO2 Entrez, Source | enolase 2 (gamma, neuronal) | 1889 | 0.112 | 0.5815 | Yes |
| 19 | MVD | MVD Entrez, Source | mevalonate (diphospho) decarboxylase | 1998 | 0.108 | 0.5868 | Yes |
| 20 | ALDOB | ALDOB Entrez, Source | aldolase B, fructose-bisphosphate | 2152 | 0.101 | 0.5879 | Yes |
| 21 | HDC | HDC Entrez, Source | histidine decarboxylase | 2155 | 0.101 | 0.6004 | Yes |
| 22 | HADHA | HADHA Entrez, Source | hydroxyacyl-Coenzyme A dehydrogenase/3-ketoacyl-Coenzyme A thiolase/enoyl-Coenzyme A hydratase (trifunctional protein), alpha subunit | 2173 | 0.101 | 0.6117 | Yes |
| 23 | ODC1 | ODC1 Entrez, Source | ornithine decarboxylase 1 | 2507 | 0.088 | 0.5975 | No |
| 24 | CA4 | CA4 Entrez, Source | carbonic anhydrase IV | 2514 | 0.087 | 0.6080 | No |
| 25 | DDC | DDC Entrez, Source | dopa decarboxylase (aromatic L-amino acid decarboxylase) | 2692 | 0.081 | 0.6047 | No |
| 26 | GUCY1A3 | GUCY1A3 Entrez, Source | guanylate cyclase 1, soluble, alpha 3 | 2816 | 0.077 | 0.6050 | No |
| 27 | ACO1 | ACO1 Entrez, Source | aconitase 1, soluble | 3462 | 0.059 | 0.5638 | No |
| 28 | ALDOC | ALDOC Entrez, Source | aldolase C, fructose-bisphosphate | 3477 | 0.058 | 0.5700 | No |
| 29 | CA5B | CA5B Entrez, Source | carbonic anhydrase VB, mitochondrial | 3787 | 0.050 | 0.5531 | No |
| 30 | SCLY | SCLY Entrez, Source | selenocysteine lyase | 3833 | 0.049 | 0.5558 | No |
| 31 | ALDOA | ALDOA Entrez, Source | aldolase A, fructose-bisphosphate | 5039 | 0.023 | 0.4681 | No |
| 32 | GUCY2F | GUCY2F Entrez, Source | guanylate cyclase 2F, retinal | 5642 | 0.012 | 0.4243 | No |
| 33 | GMDS | GMDS Entrez, Source | GDP-mannose 4,6-dehydratase | 6723 | -0.008 | 0.3440 | No |
| 34 | ASL | ASL Entrez, Source | argininosuccinate lyase | 6773 | -0.008 | 0.3413 | No |
| 35 | UROD | UROD Entrez, Source | uroporphyrinogen decarboxylase | 6930 | -0.011 | 0.3310 | No |
| 36 | OGG1 | OGG1 Entrez, Source | 8-oxoguanine DNA glycosylase | 7089 | -0.014 | 0.3208 | No |
| 37 | GLO1 | GLO1 Entrez, Source | glyoxalase I | 7131 | -0.014 | 0.3195 | No |
| 38 | ALAD | ALAD Entrez, Source | aminolevulinate, delta-, dehydratase | 7244 | -0.017 | 0.3132 | No |
| 39 | HADHB | HADHB Entrez, Source | hydroxyacyl-Coenzyme A dehydrogenase/3-ketoacyl-Coenzyme A thiolase/enoyl-Coenzyme A hydratase (trifunctional protein), beta subunit | 7978 | -0.031 | 0.2619 | No |
| 40 | GUCY1A2 | GUCY1A2 Entrez, Source | guanylate cyclase 1, soluble, alpha 2 | 8381 | -0.039 | 0.2365 | No |
| 41 | ADCY10 | 8726 | -0.046 | 0.2163 | No | ||
| 42 | GAD1 | GAD1 Entrez, Source | glutamate decarboxylase 1 (brain, 67kDa) | 9270 | -0.057 | 0.1827 | No |
| 43 | GUCY2D | GUCY2D Entrez, Source | guanylate cyclase 2D, membrane (retina-specific) | 9331 | -0.059 | 0.1855 | No |
| 44 | LTC4S | LTC4S Entrez, Source | leukotriene C4 synthase | 9342 | -0.059 | 0.1921 | No |
| 45 | UMPS | UMPS Entrez, Source | uridine monophosphate synthetase (orotate phosphoribosyl transferase and orotidine-5'-decarboxylase) | 9665 | -0.067 | 0.1763 | No |
| 46 | ADCY8 | ADCY8 Entrez, Source | adenylate cyclase 8 (brain) | 9950 | -0.075 | 0.1643 | No |
| 47 | APEX1 | APEX1 Entrez, Source | APEX nuclease (multifunctional DNA repair enzyme) 1 | 10112 | -0.079 | 0.1620 | No |
| 48 | AMD1 | AMD1 Entrez, Source | adenosylmethionine decarboxylase 1 | 10528 | -0.091 | 0.1422 | No |
| 49 | CA2 | CA2 Entrez, Source | carbonic anhydrase II | 10759 | -0.099 | 0.1372 | No |
| 50 | ENO3 | ENO3 Entrez, Source | enolase 3 (beta, muscle) | 11125 | -0.113 | 0.1239 | No |
| 51 | CA3 | CA3 Entrez, Source | carbonic anhydrase III, muscle specific | 13129 | -0.343 | 0.0160 | No |