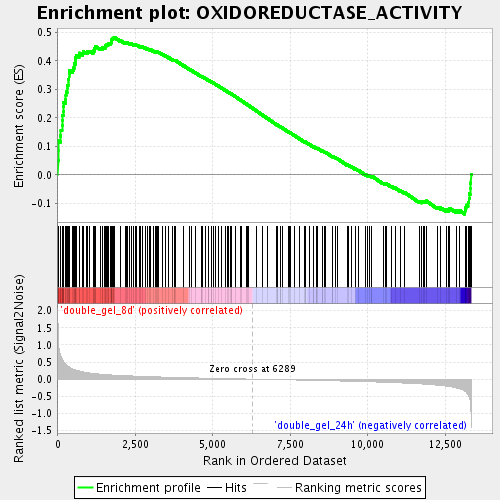
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | double_gel_and_single_gel_rma_expression_values_collapsed_to_symbols.class.cls #double_gel_8d_versus_double_gel_24h.class.cls #double_gel_8d_versus_double_gel_24h_repos |
| Phenotype | class.cls#double_gel_8d_versus_double_gel_24h_repos |
| Upregulated in class | double_gel_8d |
| GeneSet | OXIDOREDUCTASE_ACTIVITY |
| Enrichment Score (ES) | 0.48346266 |
| Normalized Enrichment Score (NES) | 1.8667878 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.016775468 |
| FWER p-Value | 0.697 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | CYP17A1 | CYP17A1 Entrez, Source | cytochrome P450, family 17, subfamily A, polypeptide 1 | 2 | 1.623 | 0.0513 | Yes |
| 2 | LDHB | LDHB Entrez, Source | lactate dehydrogenase B | 15 | 1.102 | 0.0854 | Yes |
| 3 | HAO2 | HAO2 Entrez, Source | hydroxyacid oxidase 2 (long chain) | 18 | 1.071 | 0.1192 | Yes |
| 4 | BBOX1 | BBOX1 Entrez, Source | butyrobetaine (gamma), 2-oxoglutarate dioxygenase (gamma-butyrobetaine hydroxylase) 1 | 78 | 0.695 | 0.1368 | Yes |
| 5 | ACOX2 | ACOX2 Entrez, Source | acyl-Coenzyme A oxidase 2, branched chain | 97 | 0.667 | 0.1566 | Yes |
| 6 | PAH | PAH Entrez, Source | phenylalanine hydroxylase | 133 | 0.597 | 0.1729 | Yes |
| 7 | HGD | HGD Entrez, Source | homogentisate 1,2-dioxygenase (homogentisate oxidase) | 138 | 0.591 | 0.1914 | Yes |
| 8 | MIOX | MIOX Entrez, Source | myo-inositol oxygenase | 146 | 0.570 | 0.2089 | Yes |
| 9 | TDO2 | TDO2 Entrez, Source | tryptophan 2,3-dioxygenase | 170 | 0.532 | 0.2240 | Yes |
| 10 | CYP8B1 | CYP8B1 Entrez, Source | cytochrome P450, family 8, subfamily B, polypeptide 1 | 177 | 0.522 | 0.2401 | Yes |
| 11 | ALDH1A1 | ALDH1A1 Entrez, Source | aldehyde dehydrogenase 1 family, member A1 | 185 | 0.510 | 0.2558 | Yes |
| 12 | GPX3 | GPX3 Entrez, Source | glutathione peroxidase 3 (plasma) | 242 | 0.449 | 0.2658 | Yes |
| 13 | BCKDHA | BCKDHA Entrez, Source | branched chain keto acid dehydrogenase E1, alpha polypeptide | 258 | 0.431 | 0.2783 | Yes |
| 14 | CYBA | CYBA Entrez, Source | cytochrome b-245, alpha polypeptide | 264 | 0.427 | 0.2915 | Yes |
| 15 | CYP27A1 | CYP27A1 Entrez, Source | cytochrome P450, family 27, subfamily A, polypeptide 1 | 297 | 0.403 | 0.3019 | Yes |
| 16 | ALDH6A1 | ALDH6A1 Entrez, Source | aldehyde dehydrogenase 6 family, member A1 | 300 | 0.402 | 0.3145 | Yes |
| 17 | ADH7 | ADH7 Entrez, Source | alcohol dehydrogenase 7 (class IV), mu or sigma polypeptide | 329 | 0.374 | 0.3242 | Yes |
| 18 | FADS1 | FADS1 Entrez, Source | fatty acid desaturase 1 | 330 | 0.373 | 0.3360 | Yes |
| 19 | DMGDH | DMGDH Entrez, Source | dimethylglycine dehydrogenase | 357 | 0.361 | 0.3455 | Yes |
| 20 | CYB5A | CYB5A Entrez, Source | cytochrome b5 type A (microsomal) | 359 | 0.359 | 0.3568 | Yes |
| 21 | EHHADH | EHHADH Entrez, Source | enoyl-Coenzyme A, hydratase/3-hydroxyacyl Coenzyme A dehydrogenase | 375 | 0.349 | 0.3668 | Yes |
| 22 | DECR1 | DECR1 Entrez, Source | 2,4-dienoyl CoA reductase 1, mitochondrial | 486 | 0.291 | 0.3676 | Yes |
| 23 | HSD3B1 | HSD3B1 Entrez, Source | hydroxy-delta-5-steroid dehydrogenase, 3 beta- and steroid delta-isomerase 1 | 501 | 0.286 | 0.3756 | Yes |
| 24 | POR | POR Entrez, Source | P450 (cytochrome) oxidoreductase | 523 | 0.278 | 0.3829 | Yes |
| 25 | ALDH3A2 | ALDH3A2 Entrez, Source | aldehyde dehydrogenase 3 family, member A2 | 526 | 0.277 | 0.3915 | Yes |
| 26 | LOX | LOX Entrez, Source | lysyl oxidase | 558 | 0.267 | 0.3976 | Yes |
| 27 | DHCR24 | DHCR24 Entrez, Source | 24-dehydrocholesterol reductase | 561 | 0.267 | 0.4059 | Yes |
| 28 | PHYH | PHYH Entrez, Source | phytanoyl-CoA 2-hydroxylase | 566 | 0.265 | 0.4140 | Yes |
| 29 | ALDH5A1 | ALDH5A1 Entrez, Source | aldehyde dehydrogenase 5 family, member A1 (succinate-semialdehyde dehydrogenase) | 595 | 0.258 | 0.4201 | Yes |
| 30 | ADI1 | ADI1 Entrez, Source | acireductone dioxygenase 1 | 682 | 0.239 | 0.4211 | Yes |
| 31 | CDO1 | CDO1 Entrez, Source | cysteine dioxygenase, type I | 689 | 0.237 | 0.4282 | Yes |
| 32 | IVD | IVD Entrez, Source | isovaleryl Coenzyme A dehydrogenase | 800 | 0.213 | 0.4266 | Yes |
| 33 | PHGDH | PHGDH Entrez, Source | phosphoglycerate dehydrogenase | 810 | 0.211 | 0.4326 | Yes |
| 34 | BCKDHB | BCKDHB Entrez, Source | branched chain keto acid dehydrogenase E1, beta polypeptide (maple syrup urine disease) | 906 | 0.195 | 0.4316 | Yes |
| 35 | ETFDH | ETFDH Entrez, Source | electron-transferring-flavoprotein dehydrogenase | 962 | 0.187 | 0.4333 | Yes |
| 36 | PDHA2 | PDHA2 Entrez, Source | pyruvate dehydrogenase (lipoamide) alpha 2 | 1026 | 0.179 | 0.4342 | Yes |
| 37 | IDH3G | IDH3G Entrez, Source | isocitrate dehydrogenase 3 (NAD+) gamma | 1135 | 0.167 | 0.4313 | Yes |
| 38 | SARDH | SARDH Entrez, Source | sarcosine dehydrogenase | 1150 | 0.165 | 0.4355 | Yes |
| 39 | NSDHL | NSDHL Entrez, Source | NAD(P) dependent steroid dehydrogenase-like | 1169 | 0.164 | 0.4393 | Yes |
| 40 | ALDH7A1 | ALDH7A1 Entrez, Source | aldehyde dehydrogenase 7 family, member A1 | 1179 | 0.163 | 0.4438 | Yes |
| 41 | CYB5R2 | CYB5R2 Entrez, Source | cytochrome b5 reductase 2 | 1199 | 0.161 | 0.4474 | Yes |
| 42 | RETSAT | RETSAT Entrez, Source | retinol saturase (all-trans-retinol 13,14-reductase) | 1210 | 0.160 | 0.4518 | Yes |
| 43 | MSRA | MSRA Entrez, Source | methionine sulfoxide reductase A | 1379 | 0.144 | 0.4436 | Yes |
| 44 | ALDH3A1 | ALDH3A1 Entrez, Source | aldehyde dehydrogenase 3 family, memberA1 | 1428 | 0.140 | 0.4444 | Yes |
| 45 | NOX1 | NOX1 Entrez, Source | NADPH oxidase 1 | 1443 | 0.139 | 0.4477 | Yes |
| 46 | HSD17B4 | HSD17B4 Entrez, Source | hydroxysteroid (17-beta) dehydrogenase 4 | 1489 | 0.136 | 0.4486 | Yes |
| 47 | MAOB | MAOB Entrez, Source | monoamine oxidase B | 1531 | 0.133 | 0.4497 | Yes |
| 48 | PRDX3 | PRDX3 Entrez, Source | peroxiredoxin 3 | 1534 | 0.132 | 0.4537 | Yes |
| 49 | NDUFS2 | NDUFS2 Entrez, Source | NADH dehydrogenase (ubiquinone) Fe-S protein 2, 49kDa (NADH-coenzyme Q reductase) | 1554 | 0.131 | 0.4565 | Yes |
| 50 | COX8A | COX8A Entrez, Source | cytochrome c oxidase subunit 8A (ubiquitous) | 1597 | 0.128 | 0.4574 | Yes |
| 51 | FMO2 | FMO2 Entrez, Source | flavin containing monooxygenase 2 (non-functional) | 1618 | 0.127 | 0.4599 | Yes |
| 52 | ME1 | ME1 Entrez, Source | malic enzyme 1, NADP(+)-dependent, cytosolic | 1647 | 0.125 | 0.4617 | Yes |
| 53 | HPGD | HPGD Entrez, Source | hydroxyprostaglandin dehydrogenase 15-(NAD) | 1689 | 0.123 | 0.4625 | Yes |
| 54 | ACADSB | ACADSB Entrez, Source | acyl-Coenzyme A dehydrogenase, short/branched chain | 1715 | 0.122 | 0.4645 | Yes |
| 55 | ACADL | ACADL Entrez, Source | acyl-Coenzyme A dehydrogenase, long chain | 1720 | 0.122 | 0.4680 | Yes |
| 56 | SUOX | SUOX Entrez, Source | sulfite oxidase | 1734 | 0.121 | 0.4709 | Yes |
| 57 | ALOX5 | ALOX5 Entrez, Source | arachidonate 5-lipoxygenase | 1736 | 0.120 | 0.4746 | Yes |
| 58 | COX7B | COX7B Entrez, Source | cytochrome c oxidase subunit VIIb | 1747 | 0.119 | 0.4777 | Yes |
| 59 | DHRS9 | DHRS9 Entrez, Source | dehydrogenase/reductase (SDR family) member 9 | 1778 | 0.118 | 0.4791 | Yes |
| 60 | SCD | SCD Entrez, Source | stearoyl-CoA desaturase (delta-9-desaturase) | 1803 | 0.117 | 0.4810 | Yes |
| 61 | HMOX2 | HMOX2 Entrez, Source | heme oxygenase (decycling) 2 | 1820 | 0.116 | 0.4835 | Yes |
| 62 | HAAO | HAAO Entrez, Source | 3-hydroxyanthranilate 3,4-dioxygenase | 2017 | 0.107 | 0.4720 | No |
| 63 | HADHA | HADHA Entrez, Source | hydroxyacyl-Coenzyme A dehydrogenase/3-ketoacyl-Coenzyme A thiolase/enoyl-Coenzyme A hydratase (trifunctional protein), alpha subunit | 2173 | 0.101 | 0.4634 | No |
| 64 | ALOX15 | ALOX15 Entrez, Source | arachidonate 15-lipoxygenase | 2217 | 0.099 | 0.4632 | No |
| 65 | CYB5R4 | CYB5R4 Entrez, Source | cytochrome b5 reductase 4 | 2233 | 0.098 | 0.4652 | No |
| 66 | COX4I2 | COX4I2 Entrez, Source | cytochrome c oxidase subunit IV isoform 2 (lung) | 2312 | 0.095 | 0.4623 | No |
| 67 | TXN2 | TXN2 Entrez, Source | thioredoxin 2 | 2370 | 0.093 | 0.4609 | No |
| 68 | EGLN2 | EGLN2 Entrez, Source | egl nine homolog 2 (C. elegans) | 2444 | 0.090 | 0.4582 | No |
| 69 | UQCRH | UQCRH Entrez, Source | ubiquinol-cytochrome c reductase hinge protein | 2494 | 0.088 | 0.4573 | No |
| 70 | ALDH2 | ALDH2 Entrez, Source | aldehyde dehydrogenase 2 family (mitochondrial) | 2544 | 0.086 | 0.4563 | No |
| 71 | CYP19A1 | CYP19A1 Entrez, Source | cytochrome P450, family 19, subfamily A, polypeptide 1 | 2640 | 0.082 | 0.4516 | No |
| 72 | MDH1 | MDH1 Entrez, Source | malate dehydrogenase 1, NAD (soluble) | 2674 | 0.081 | 0.4517 | No |
| 73 | GPX4 | GPX4 Entrez, Source | glutathione peroxidase 4 (phospholipid hydroperoxidase) | 2713 | 0.080 | 0.4513 | No |
| 74 | SDHD | SDHD Entrez, Source | succinate dehydrogenase complex, subunit D, integral membrane protein | 2818 | 0.077 | 0.4459 | No |
| 75 | ALDH9A1 | ALDH9A1 Entrez, Source | aldehyde dehydrogenase 9 family, member A1 | 2893 | 0.074 | 0.4426 | No |
| 76 | NOS3 | NOS3 Entrez, Source | nitric oxide synthase 3 (endothelial cell) | 2951 | 0.072 | 0.4406 | No |
| 77 | ACADVL | ACADVL Entrez, Source | acyl-Coenzyme A dehydrogenase, very long chain | 3002 | 0.071 | 0.4390 | No |
| 78 | HADH | HADH Entrez, Source | hydroxyacyl-Coenzyme A dehydrogenase | 3083 | 0.069 | 0.4351 | No |
| 79 | IDH1 | IDH1 Entrez, Source | isocitrate dehydrogenase 1 (NADP+), soluble | 3161 | 0.067 | 0.4314 | No |
| 80 | CYGB | CYGB Entrez, Source | cytoglobin | 3172 | 0.067 | 0.4328 | No |
| 81 | ETFA | ETFA Entrez, Source | electron-transfer-flavoprotein, alpha polypeptide (glutaric aciduria II) | 3209 | 0.065 | 0.4321 | No |
| 82 | CYCS | CYCS Entrez, Source | cytochrome c, somatic | 3256 | 0.064 | 0.4306 | No |
| 83 | MSRB2 | MSRB2 Entrez, Source | methionine sulfoxide reductase B2 | 3376 | 0.061 | 0.4235 | No |
| 84 | AKR7A2 | AKR7A2 Entrez, Source | aldo-keto reductase family 7, member A2 (aflatoxin aldehyde reductase) | 3458 | 0.059 | 0.4192 | No |
| 85 | DEGS1 | DEGS1 Entrez, Source | degenerative spermatocyte homolog 1, lipid desaturase (Drosophila) | 3581 | 0.055 | 0.4117 | No |
| 86 | ALDH1A2 | ALDH1A2 Entrez, Source | aldehyde dehydrogenase 1 family, member A2 | 3710 | 0.052 | 0.4036 | No |
| 87 | PDHB | PDHB Entrez, Source | pyruvate dehydrogenase (lipoamide) beta | 3755 | 0.051 | 0.4019 | No |
| 88 | ABP1 | ABP1 Entrez, Source | amiloride binding protein 1 (amine oxidase (copper-containing)) | 3774 | 0.051 | 0.4021 | No |
| 89 | ACADM | ACADM Entrez, Source | acyl-Coenzyme A dehydrogenase, C-4 to C-12 straight chain | 3794 | 0.050 | 0.4023 | No |
| 90 | PRDX2 | PRDX2 Entrez, Source | peroxiredoxin 2 | 4063 | 0.043 | 0.3833 | No |
| 91 | SDHA | SDHA Entrez, Source | succinate dehydrogenase complex, subunit A, flavoprotein (Fp) | 4257 | 0.038 | 0.3698 | No |
| 92 | GLUD1 | GLUD1 Entrez, Source | glutamate dehydrogenase 1 | 4297 | 0.038 | 0.3681 | No |
| 93 | MECR | MECR Entrez, Source | mitochondrial trans-2-enoyl-CoA reductase | 4444 | 0.035 | 0.3581 | No |
| 94 | ADH6 | ADH6 Entrez, Source | alcohol dehydrogenase 6 (class V) | 4624 | 0.032 | 0.3455 | No |
| 95 | SC5DL | SC5DL Entrez, Source | sterol-C5-desaturase (ERG3 delta-5-desaturase homolog, fungal)-like | 4644 | 0.031 | 0.3450 | No |
| 96 | COX5A | COX5A Entrez, Source | cytochrome c oxidase subunit Va | 4672 | 0.031 | 0.3439 | No |
| 97 | NDUFS4 | NDUFS4 Entrez, Source | NADH dehydrogenase (ubiquinone) Fe-S protein 4, 18kDa (NADH-coenzyme Q reductase) | 4767 | 0.029 | 0.3377 | No |
| 98 | NDUFA9 | NDUFA9 Entrez, Source | NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 9, 39kDa | 4853 | 0.027 | 0.3321 | No |
| 99 | GLRX2 | GLRX2 Entrez, Source | glutaredoxin 2 | 4870 | 0.027 | 0.3317 | No |
| 100 | PRDX4 | PRDX4 Entrez, Source | peroxiredoxin 4 | 4953 | 0.025 | 0.3263 | No |
| 101 | ETFB | ETFB Entrez, Source | electron-transfer-flavoprotein, beta polypeptide | 4956 | 0.025 | 0.3269 | No |
| 102 | RDH10 | RDH10 Entrez, Source | retinol dehydrogenase 10 (all-trans) | 5029 | 0.023 | 0.3222 | No |
| 103 | CP | CP Entrez, Source | ceruloplasmin (ferroxidase) | 5093 | 0.022 | 0.3181 | No |
| 104 | CYBRD1 | CYBRD1 Entrez, Source | cytochrome b reductase 1 | 5173 | 0.021 | 0.3128 | No |
| 105 | NOS1 | NOS1 Entrez, Source | nitric oxide synthase 1 (neuronal) | 5282 | 0.019 | 0.3052 | No |
| 106 | COX5B | COX5B Entrez, Source | cytochrome c oxidase subunit Vb | 5409 | 0.017 | 0.2961 | No |
| 107 | NOX4 | NOX4 Entrez, Source | NADPH oxidase 4 | 5475 | 0.015 | 0.2917 | No |
| 108 | MTRR | MTRR Entrez, Source | 5-methyltetrahydrofolate-homocysteine methyltransferase reductase | 5503 | 0.015 | 0.2901 | No |
| 109 | PTGS1 | PTGS1 Entrez, Source | prostaglandin-endoperoxide synthase 1 (prostaglandin G/H synthase and cyclooxygenase) | 5556 | 0.014 | 0.2866 | No |
| 110 | AOC3 | AOC3 Entrez, Source | amine oxidase, copper containing 3 (vascular adhesion protein 1) | 5594 | 0.013 | 0.2841 | No |
| 111 | COX4I1 | COX4I1 Entrez, Source | cytochrome c oxidase subunit IV isoform 1 | 5610 | 0.013 | 0.2834 | No |
| 112 | DHCR7 | DHCR7 Entrez, Source | 7-dehydrocholesterol reductase | 5728 | 0.010 | 0.2748 | No |
| 113 | SC4MOL | SC4MOL Entrez, Source | sterol-C4-methyl oxidase-like | 5880 | 0.008 | 0.2636 | No |
| 114 | UQCRC1 | UQCRC1 Entrez, Source | ubiquinol-cytochrome c reductase core protein I | 5927 | 0.007 | 0.2603 | No |
| 115 | DLD | DLD Entrez, Source | dihydrolipoamide dehydrogenase | 6088 | 0.003 | 0.2483 | No |
| 116 | SRD5A2 | SRD5A2 Entrez, Source | steroid-5-alpha-reductase, alpha polypeptide 2 (3-oxo-5 alpha-steroid delta 4-dehydrogenase alpha 2) | 6110 | 0.003 | 0.2468 | No |
| 117 | NCF1 | NCF1 Entrez, Source | neutrophil cytosolic factor 1, (chronic granulomatous disease, autosomal 1) | 6138 | 0.003 | 0.2448 | No |
| 118 | GPX2 | GPX2 Entrez, Source | glutathione peroxidase 2 (gastrointestinal) | 6396 | -0.002 | 0.2253 | No |
| 119 | ADH4 | ADH4 Entrez, Source | alcohol dehydrogenase 4 (class II), pi polypeptide | 6589 | -0.005 | 0.2109 | No |
| 120 | PRODH | PRODH Entrez, Source | proline dehydrogenase (oxidase) 1 | 6759 | -0.008 | 0.1983 | No |
| 121 | CYP7B1 | CYP7B1 Entrez, Source | cytochrome P450, family 7, subfamily B, polypeptide 1 | 7069 | -0.014 | 0.1753 | No |
| 122 | XDH | XDH Entrez, Source | xanthine dehydrogenase | 7075 | -0.014 | 0.1753 | No |
| 123 | SOD1 | SOD1 Entrez, Source | superoxide dismutase 1, soluble (amyotrophic lateral sclerosis 1 (adult)) | 7093 | -0.014 | 0.1744 | No |
| 124 | CPOX | CPOX Entrez, Source | coproporphyrinogen oxidase | 7172 | -0.015 | 0.1690 | No |
| 125 | FDXR | FDXR Entrez, Source | ferredoxin reductase | 7183 | -0.015 | 0.1687 | No |
| 126 | CYP1B1 | CYP1B1 Entrez, Source | cytochrome P450, family 1, subfamily B, polypeptide 1 | 7236 | -0.017 | 0.1653 | No |
| 127 | QDPR | QDPR Entrez, Source | quinoid dihydropteridine reductase | 7441 | -0.021 | 0.1505 | No |
| 128 | TH | TH Entrez, Source | tyrosine hydroxylase | 7473 | -0.021 | 0.1488 | No |
| 129 | RDH11 | RDH11 Entrez, Source | retinol dehydrogenase 11 (all-trans/9-cis/11-cis) | 7495 | -0.022 | 0.1479 | No |
| 130 | GAPDHS | GAPDHS Entrez, Source | glyceraldehyde-3-phosphate dehydrogenase, spermatogenic | 7619 | -0.024 | 0.1393 | No |
| 131 | EGLN1 | EGLN1 Entrez, Source | egl nine homolog 1 (C. elegans) | 7809 | -0.028 | 0.1258 | No |
| 132 | NDUFS7 | NDUFS7 Entrez, Source | NADH dehydrogenase (ubiquinone) Fe-S protein 7, 20kDa (NADH-coenzyme Q reductase) | 7963 | -0.031 | 0.1151 | No |
| 133 | TXNL1 | TXNL1 Entrez, Source | thioredoxin-like 1 | 7977 | -0.031 | 0.1151 | No |
| 134 | HADHB | HADHB Entrez, Source | hydroxyacyl-Coenzyme A dehydrogenase/3-ketoacyl-Coenzyme A thiolase/enoyl-Coenzyme A hydratase (trifunctional protein), beta subunit | 7978 | -0.031 | 0.1161 | No |
| 135 | CYP4F2 | CYP4F2 Entrez, Source | cytochrome P450, family 4, subfamily F, polypeptide 2 | 8129 | -0.034 | 0.1058 | No |
| 136 | COX15 | COX15 Entrez, Source | COX15 homolog, cytochrome c oxidase assembly protein (yeast) | 8234 | -0.036 | 0.0990 | No |
| 137 | NDUFS1 | NDUFS1 Entrez, Source | NADH dehydrogenase (ubiquinone) Fe-S protein 1, 75kDa (NADH-coenzyme Q reductase) | 8263 | -0.037 | 0.0980 | No |
| 138 | NQO2 | NQO2 Entrez, Source | NAD(P)H dehydrogenase, quinone 2 | 8329 | -0.038 | 0.0943 | No |
| 139 | AKR1B10 | AKR1B10 Entrez, Source | aldo-keto reductase family 1, member B10 (aldose reductase) | 8361 | -0.039 | 0.0932 | No |
| 140 | GMPR | GMPR Entrez, Source | guanosine monophosphate reductase | 8380 | -0.039 | 0.0930 | No |
| 141 | CYB5R3 | CYB5R3 Entrez, Source | cytochrome b5 reductase 3 | 8531 | -0.042 | 0.0830 | No |
| 142 | DIO2 | DIO2 Entrez, Source | deiodinase, iodothyronine, type II | 8533 | -0.042 | 0.0842 | No |
| 143 | TXNDC1 | TXNDC1 Entrez, Source | thioredoxin domain containing 1 | 8611 | -0.044 | 0.0798 | No |
| 144 | GPD1 | GPD1 Entrez, Source | glycerol-3-phosphate dehydrogenase 1 (soluble) | 8635 | -0.044 | 0.0794 | No |
| 145 | IDH3B | IDH3B Entrez, Source | isocitrate dehydrogenase 3 (NAD+) beta | 8861 | -0.049 | 0.0639 | No |
| 146 | CAT | CAT Entrez, Source | catalase | 8873 | -0.049 | 0.0646 | No |
| 147 | AKR1A1 | AKR1A1 Entrez, Source | aldo-keto reductase family 1, member A1 (aldehyde reductase) | 8947 | -0.051 | 0.0607 | No |
| 148 | CYP3A4 | CYP3A4 Entrez, Source | cytochrome P450, family 3, subfamily A, polypeptide 4 | 9007 | -0.052 | 0.0578 | No |
| 149 | NQO1 | NQO1 Entrez, Source | NAD(P)H dehydrogenase, quinone 1 | 9332 | -0.059 | 0.0351 | No |
| 150 | BLVRA | BLVRA Entrez, Source | biliverdin reductase A | 9379 | -0.060 | 0.0335 | No |
| 151 | PLOD3 | PLOD3 Entrez, Source | procollagen-lysine, 2-oxoglutarate 5-dioxygenase 3 | 9471 | -0.062 | 0.0285 | No |
| 152 | SPR | SPR Entrez, Source | sepiapterin reductase (7,8-dihydrobiopterin:NADP+ oxidoreductase) | 9602 | -0.065 | 0.0207 | No |
| 153 | HSD17B7 | HSD17B7 Entrez, Source | hydroxysteroid (17-beta) dehydrogenase 7 | 9685 | -0.067 | 0.0166 | No |
| 154 | LDHA | LDHA Entrez, Source | lactate dehydrogenase A | 9934 | -0.075 | 0.0002 | No |
| 155 | LDHC | LDHC Entrez, Source | lactate dehydrogenase C | 10001 | -0.077 | -0.0024 | No |
| 156 | CRYZL1 | CRYZL1 Entrez, Source | crystallin, zeta (quinone reductase)-like 1 | 10057 | -0.078 | -0.0041 | No |
| 157 | APEX1 | APEX1 Entrez, Source | APEX nuclease (multifunctional DNA repair enzyme) 1 | 10112 | -0.079 | -0.0057 | No |
| 158 | P4HB | P4HB Entrez, Source | procollagen-proline, 2-oxoglutarate 4-dioxygenase (proline 4-hydroxylase), beta polypeptide | 10130 | -0.080 | -0.0045 | No |
| 159 | DUOX2 | DUOX2 Entrez, Source | dual oxidase 2 | 10498 | -0.090 | -0.0295 | No |
| 160 | INDO | INDO Entrez, Source | indoleamine-pyrrole 2,3 dioxygenase | 10558 | -0.092 | -0.0311 | No |
| 161 | TXNRD2 | TXNRD2 Entrez, Source | thioredoxin reductase 2 | 10598 | -0.093 | -0.0311 | No |
| 162 | CYP11B1 | CYP11B1 Entrez, Source | cytochrome P450, family 11, subfamily B, polypeptide 1 | 10757 | -0.099 | -0.0399 | No |
| 163 | ACOX3 | ACOX3 Entrez, Source | acyl-Coenzyme A oxidase 3, pristanoyl | 10884 | -0.104 | -0.0462 | No |
| 164 | ALDH3B1 | ALDH3B1 Entrez, Source | aldehyde dehydrogenase 3 family, member B1 | 11040 | -0.109 | -0.0545 | No |
| 165 | CBR1 | CBR1 Entrez, Source | carbonyl reductase 1 | 11177 | -0.115 | -0.0612 | No |
| 166 | PDIA5 | PDIA5 Entrez, Source | protein disulfide isomerase family A, member 5 | 11658 | -0.136 | -0.0934 | No |
| 167 | P4HA1 | P4HA1 Entrez, Source | procollagen-proline, 2-oxoglutarate 4-dioxygenase (proline 4-hydroxylase), alpha polypeptide I | 11736 | -0.140 | -0.0948 | No |
| 168 | FDX1 | FDX1 Entrez, Source | ferredoxin 1 | 11784 | -0.144 | -0.0938 | No |
| 169 | NUDT5 | NUDT5 Entrez, Source | nudix (nucleoside diphosphate linked moiety X)-type motif 5 | 11827 | -0.146 | -0.0923 | No |
| 170 | SOD2 | SOD2 Entrez, Source | superoxide dismutase 2, mitochondrial | 11883 | -0.149 | -0.0918 | No |
| 171 | SURF1 | SURF1 Entrez, Source | surfeit 1 | 12247 | -0.175 | -0.1138 | No |
| 172 | KMO | KMO Entrez, Source | kynurenine 3-monooxygenase (kynurenine 3-hydroxylase) | 12348 | -0.183 | -0.1156 | No |
| 173 | DHRS3 | DHRS3 Entrez, Source | dehydrogenase/reductase (SDR family) member 3 | 12528 | -0.202 | -0.1228 | No |
| 174 | IMPDH2 | IMPDH2 Entrez, Source | IMP (inosine monophosphate) dehydrogenase 2 | 12613 | -0.213 | -0.1224 | No |
| 175 | GPD2 | GPD2 Entrez, Source | glycerol-3-phosphate dehydrogenase 2 (mitochondrial) | 12638 | -0.216 | -0.1174 | No |
| 176 | FMO1 | FMO1 Entrez, Source | flavin containing monooxygenase 1 | 12848 | -0.253 | -0.1252 | No |
| 177 | TXNRD1 | TXNRD1 Entrez, Source | thioredoxin reductase 1 | 12967 | -0.282 | -0.1253 | No |
| 178 | UGDH | UGDH Entrez, Source | UDP-glucose dehydrogenase | 13138 | -0.352 | -0.1270 | No |
| 179 | SRD5A1 | SRD5A1 Entrez, Source | steroid-5-alpha-reductase, alpha polypeptide 1 (3-oxo-5 alpha-steroid delta 4-dehydrogenase alpha 1) | 13143 | -0.354 | -0.1161 | No |
| 180 | GSR | GSR Entrez, Source | glutathione reductase | 13171 | -0.376 | -0.1062 | No |
| 181 | AKR7A3 | AKR7A3 Entrez, Source | aldo-keto reductase family 7, member A3 (aflatoxin aldehyde reductase) | 13236 | -0.447 | -0.0969 | No |
| 182 | HMOX1 | HMOX1 Entrez, Source | heme oxygenase (decycling) 1 | 13267 | -0.519 | -0.0827 | No |
| 183 | CYP1A2 | CYP1A2 Entrez, Source | cytochrome P450, family 1, subfamily A, polypeptide 2 | 13275 | -0.526 | -0.0665 | No |
| 184 | AOX1 | AOX1 Entrez, Source | aldehyde oxidase 1 | 13304 | -0.615 | -0.0492 | No |
| 185 | BDH1 | BDH1 Entrez, Source | 3-hydroxybutyrate dehydrogenase, type 1 | 13310 | -0.654 | -0.0288 | No |
| 186 | CYP26A1 | CYP26A1 Entrez, Source | cytochrome P450, family 26, subfamily A, polypeptide 1 | 13332 | -0.982 | 0.0008 | No |

