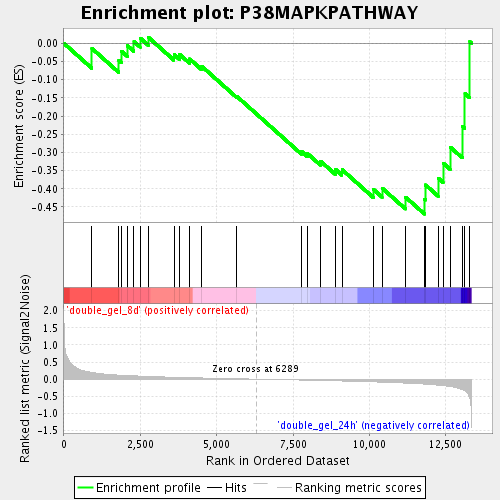
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | double_gel_and_single_gel_rma_expression_values_collapsed_to_symbols.class.cls #double_gel_8d_versus_double_gel_24h.class.cls #double_gel_8d_versus_double_gel_24h_repos |
| Phenotype | class.cls#double_gel_8d_versus_double_gel_24h_repos |
| Upregulated in class | double_gel_24h |
| GeneSet | P38MAPKPATHWAY |
| Enrichment Score (ES) | -0.4696239 |
| Normalized Enrichment Score (NES) | -1.4037218 |
| Nominal p-value | 0.0625 |
| FDR q-value | 0.29234418 |
| FWER p-Value | 1.0 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | TGFB2 | TGFB2 Entrez, Source | transforming growth factor, beta 2 | 916 | 0.193 | -0.0140 | No |
| 2 | DAXX | DAXX Entrez, Source | death-associated protein 6 | 1796 | 0.117 | -0.0469 | No |
| 3 | MAPKAPK2 | MAPKAPK2 Entrez, Source | mitogen-activated protein kinase-activated protein kinase 2 | 1883 | 0.113 | -0.0214 | No |
| 4 | TGFB1 | TGFB1 Entrez, Source | transforming growth factor, beta 1 (Camurati-Engelmann disease) | 2076 | 0.105 | -0.0062 | No |
| 5 | HSPB2 | HSPB2 Entrez, Source | heat shock 27kDa protein 2 | 2292 | 0.096 | 0.0048 | No |
| 6 | MAX | MAX Entrez, Source | MYC associated factor X | 2495 | 0.088 | 0.0146 | No |
| 7 | HRAS | HRAS Entrez, Source | v-Ha-ras Harvey rat sarcoma viral oncogene homolog | 2761 | 0.078 | 0.0167 | No |
| 8 | MAPK14 | MAPK14 Entrez, Source | mitogen-activated protein kinase 14 | 3602 | 0.055 | -0.0308 | No |
| 9 | PLA2G4A | PLA2G4A Entrez, Source | phospholipase A2, group IVA (cytosolic, calcium-dependent) | 3785 | 0.050 | -0.0302 | No |
| 10 | TGFB3 | TGFB3 Entrez, Source | transforming growth factor, beta 3 | 4113 | 0.042 | -0.0429 | No |
| 11 | MAP2K6 | MAP2K6 Entrez, Source | mitogen-activated protein kinase kinase 6 | 4510 | 0.034 | -0.0631 | No |
| 12 | TRADD | TRADD Entrez, Source | TNFRSF1A-associated via death domain | 5653 | 0.012 | -0.1455 | No |
| 13 | SHC1 | SHC1 Entrez, Source | SHC (Src homology 2 domain containing) transforming protein 1 | 7776 | -0.027 | -0.2972 | No |
| 14 | MAP2K4 | MAP2K4 Entrez, Source | mitogen-activated protein kinase kinase 4 | 7969 | -0.031 | -0.3030 | No |
| 15 | ELK1 | ELK1 Entrez, Source | ELK1, member of ETS oncogene family | 8398 | -0.039 | -0.3240 | No |
| 16 | CDC42 | CDC42 Entrez, Source | cell division cycle 42 (GTP binding protein, 25kDa) | 8893 | -0.050 | -0.3470 | No |
| 17 | RAC1 | RAC1 Entrez, Source | ras-related C3 botulinum toxin substrate 1 (rho family, small GTP binding protein Rac1) | 9101 | -0.054 | -0.3473 | No |
| 18 | HMGN1 | HMGN1 Entrez, Source | high-mobility group nucleosome binding domain 1 | 10136 | -0.080 | -0.4022 | No |
| 19 | MEF2D | MEF2D Entrez, Source | MADS box transcription enhancer factor 2, polypeptide D (myocyte enhancer factor 2D) | 10414 | -0.088 | -0.3982 | No |
| 20 | STAT1 | STAT1 Entrez, Source | signal transducer and activator of transcription 1, 91kDa | 11180 | -0.115 | -0.4229 | No |
| 21 | GRB2 | GRB2 Entrez, Source | growth factor receptor-bound protein 2 | 11803 | -0.145 | -0.4286 | Yes |
| 22 | TGFBR1 | TGFBR1 Entrez, Source | transforming growth factor, beta receptor I (activin A receptor type II-like kinase, 53kDa) | 11835 | -0.147 | -0.3894 | Yes |
| 23 | MYC | MYC Entrez, Source | v-myc myelocytomatosis viral oncogene homolog (avian) | 12249 | -0.175 | -0.3707 | Yes |
| 24 | MAP3K1 | MAP3K1 Entrez, Source | mitogen-activated protein kinase kinase kinase 1 | 12410 | -0.189 | -0.3292 | Yes |
| 25 | ATF2 | ATF2 Entrez, Source | activating transcription factor 2 | 12642 | -0.216 | -0.2853 | Yes |
| 26 | CREB1 | CREB1 Entrez, Source | cAMP responsive element binding protein 1 | 13043 | -0.306 | -0.2286 | Yes |
| 27 | HSPB1 | HSPB1 Entrez, Source | heat shock 27kDa protein 1 | 13125 | -0.341 | -0.1381 | Yes |
| 28 | DDIT3 | DDIT3 Entrez, Source | DNA-damage-inducible transcript 3 | 13285 | -0.545 | 0.0043 | Yes |

