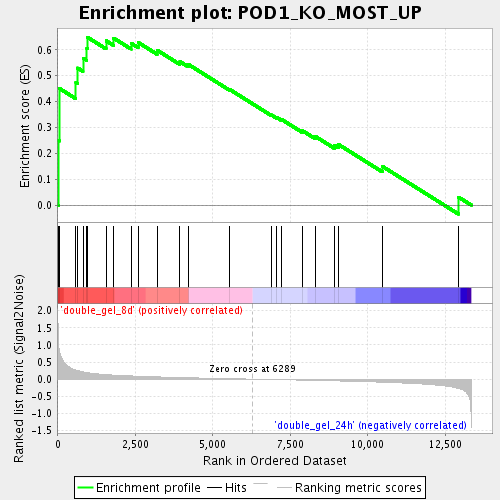
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | double_gel_and_single_gel_rma_expression_values_collapsed_to_symbols.class.cls #double_gel_8d_versus_double_gel_24h.class.cls #double_gel_8d_versus_double_gel_24h_repos |
| Phenotype | class.cls#double_gel_8d_versus_double_gel_24h_repos |
| Upregulated in class | double_gel_8d |
| GeneSet | POD1_KO_MOST_UP |
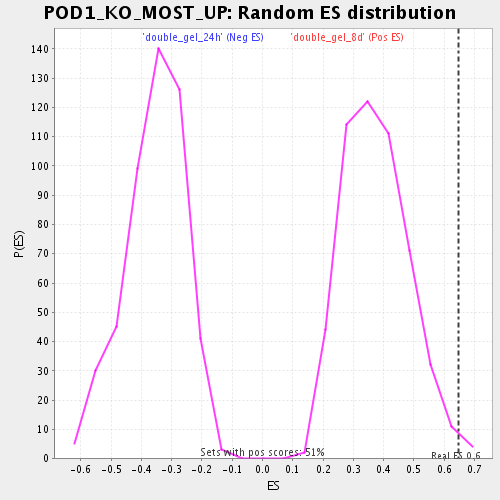
| Enrichment Score (ES) | 0.6484023 |
| Normalized Enrichment Score (NES) | 1.7272171 |
| Nominal p-value | 0.011741683 |
| FDR q-value | 0.04684414 |
| FWER p-Value | 1.0 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | CCND2 | CCND2 Entrez, Source | cyclin D2 | 21 | 1.039 | 0.2491 | Yes |
| 2 | COL3A1 | COL3A1 Entrez, Source | collagen, type III, alpha 1 (Ehlers-Danlos syndrome type IV, autosomal dominant) | 43 | 0.843 | 0.4511 | Yes |
| 3 | COL6A2 | COL6A2 Entrez, Source | collagen, type VI, alpha 2 | 579 | 0.262 | 0.4742 | Yes |
| 4 | LBP | LBP Entrez, Source | lipopolysaccharide binding protein | 636 | 0.247 | 0.5297 | Yes |
| 5 | PHGDH | PHGDH Entrez, Source | phosphoglycerate dehydrogenase | 810 | 0.211 | 0.5677 | Yes |
| 6 | IGSF4 | IGSF4 Entrez, Source | immunoglobulin superfamily, member 4 | 932 | 0.190 | 0.6046 | Yes |
| 7 | STARD10 | STARD10 Entrez, Source | START domain containing 10 | 953 | 0.188 | 0.6484 | Yes |
| 8 | IGFBP2 | IGFBP2 Entrez, Source | insulin-like growth factor binding protein 2, 36kDa | 1556 | 0.131 | 0.6349 | No |
| 9 | CDH6 | CDH6 Entrez, Source | cadherin 6, type 2, K-cadherin (fetal kidney) | 1797 | 0.117 | 0.6451 | No |
| 10 | SERPINF1 | SERPINF1 Entrez, Source | serpin peptidase inhibitor, clade F (alpha-2 antiplasmin, pigment epithelium derived factor), member 1 | 2361 | 0.093 | 0.6253 | No |
| 11 | RBP1 | RBP1 Entrez, Source | retinol binding protein 1, cellular | 2591 | 0.084 | 0.6283 | No |
| 12 | CDKN1B | CDKN1B Entrez, Source | cyclin-dependent kinase inhibitor 1B (p27, Kip1) | 3202 | 0.066 | 0.5984 | No |
| 13 | HMMR | HMMR Entrez, Source | hyaluronan-mediated motility receptor (RHAMM) | 3937 | 0.046 | 0.5544 | No |
| 14 | CFH | CFH Entrez, Source | complement factor H | 4209 | 0.039 | 0.5436 | No |
| 15 | SOX11 | SOX11 Entrez, Source | SRY (sex determining region Y)-box 11 | 5542 | 0.014 | 0.4469 | No |
| 16 | NCAM1 | NCAM1 Entrez, Source | neural cell adhesion molecule 1 | 6887 | -0.010 | 0.3485 | No |
| 17 | CYP7B1 | CYP7B1 Entrez, Source | cytochrome P450, family 7, subfamily B, polypeptide 1 | 7069 | -0.014 | 0.3381 | No |
| 18 | DCN | DCN Entrez, Source | decorin | 7217 | -0.016 | 0.3311 | No |
| 19 | CSPG2 | CSPG2 Entrez, Source | chondroitin sulfate proteoglycan 2 (versican) | 7897 | -0.029 | 0.2871 | No |
| 20 | MDK | MDK Entrez, Source | midkine (neurite growth-promoting factor 2) | 8318 | -0.038 | 0.2647 | No |
| 21 | LUM | LUM Entrez, Source | lumican | 8919 | -0.050 | 0.2318 | No |
| 22 | PTN | PTN Entrez, Source | pleiotrophin (heparin binding growth factor 8, neurite growth-promoting factor 1) | 9055 | -0.053 | 0.2344 | No |
| 23 | FAP | FAP Entrez, Source | fibroblast activation protein, alpha | 10470 | -0.089 | 0.1499 | No |
| 24 | HOMER2 | HOMER2 Entrez, Source | homer homolog 2 (Drosophila) | 12928 | -0.272 | 0.0311 | No |