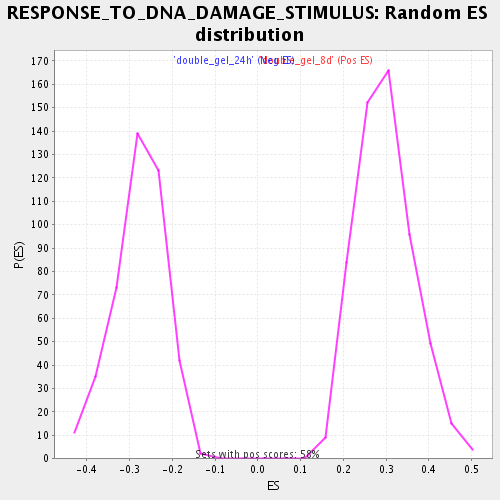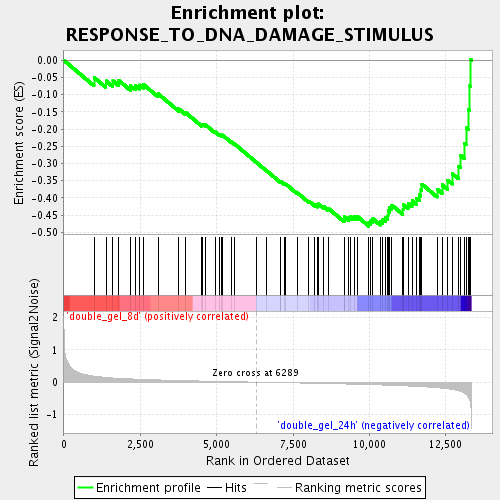
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | double_gel_and_single_gel_rma_expression_values_collapsed_to_symbols.class.cls #double_gel_8d_versus_double_gel_24h.class.cls #double_gel_8d_versus_double_gel_24h_repos |
| Phenotype | class.cls#double_gel_8d_versus_double_gel_24h_repos |
| Upregulated in class | double_gel_24h |
| GeneSet | RESPONSE_TO_DNA_DAMAGE_STIMULUS |
| Enrichment Score (ES) | -0.48260093 |
| Normalized Enrichment Score (NES) | -1.7359625 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.07858944 |
| FWER p-Value | 0.979 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | POLG | POLG Entrez, Source | polymerase (DNA directed), gamma | 985 | 0.185 | -0.0496 | No |
| 2 | MSH3 | MSH3 Entrez, Source | mutS homolog 3 (E. coli) | 1377 | 0.144 | -0.0598 | No |
| 3 | CIB1 | CIB1 Entrez, Source | calcium and integrin binding 1 (calmyrin) | 1605 | 0.128 | -0.0599 | No |
| 4 | MAPK12 | MAPK12 Entrez, Source | mitogen-activated protein kinase 12 | 1793 | 0.117 | -0.0584 | No |
| 5 | CHEK2 | CHEK2 Entrez, Source | CHK2 checkpoint homolog (S. pombe) | 2183 | 0.100 | -0.0743 | No |
| 6 | HMGB1 | HMGB1 Entrez, Source | high-mobility group box 1 | 2337 | 0.094 | -0.0734 | No |
| 7 | PARP1 | PARP1 Entrez, Source | poly (ADP-ribose) polymerase family, member 1 | 2474 | 0.089 | -0.0717 | No |
| 8 | XRCC4 | XRCC4 Entrez, Source | X-ray repair complementing defective repair in Chinese hamster cells 4 | 2599 | 0.083 | -0.0700 | No |
| 9 | XRCC6 | XRCC6 Entrez, Source | X-ray repair complementing defective repair in Chinese hamster cells 6 (Ku autoantigen, 70kDa) | 3080 | 0.069 | -0.0969 | No |
| 10 | BTG2 | BTG2 Entrez, Source | BTG family, member 2 | 3759 | 0.051 | -0.1412 | No |
| 11 | UBE2B | UBE2B Entrez, Source | ubiquitin-conjugating enzyme E2B (RAD6 homolog) | 3964 | 0.045 | -0.1505 | No |
| 12 | MAP2K6 | MAP2K6 Entrez, Source | mitogen-activated protein kinase kinase 6 | 4510 | 0.034 | -0.1871 | No |
| 13 | ATRX | ATRX Entrez, Source | alpha thalassemia/mental retardation syndrome X-linked (RAD54 homolog, S. cerevisiae) | 4548 | 0.033 | -0.1855 | No |
| 14 | BRE | BRE Entrez, Source | brain and reproductive organ-expressed (TNFRSF1A modulator) | 4635 | 0.031 | -0.1878 | No |
| 15 | RAD23A | RAD23A Entrez, Source | RAD23 homolog A (S. cerevisiae) | 4946 | 0.025 | -0.2078 | No |
| 16 | SPDYA | SPDYA Entrez, Source | speedy homolog A (Drosophila) | 5077 | 0.023 | -0.2146 | No |
| 17 | MSH2 | MSH2 Entrez, Source | mutS homolog 2, colon cancer, nonpolyposis type 1 (E. coli) | 5162 | 0.021 | -0.2181 | No |
| 18 | CCNA2 | CCNA2 Entrez, Source | cyclin A2 | 5186 | 0.020 | -0.2171 | No |
| 19 | SUMO1 | SUMO1 Entrez, Source | SMT3 suppressor of mif two 3 homolog 1 (S. cerevisiae) | 5474 | 0.015 | -0.2367 | No |
| 20 | UBE2N | UBE2N Entrez, Source | ubiquitin-conjugating enzyme E2N (UBC13 homolog, yeast) | 5570 | 0.013 | -0.2421 | No |
| 21 | RECQL | RECQL Entrez, Source | RecQ protein-like (DNA helicase Q1-like) | 6311 | -0.000 | -0.2978 | No |
| 22 | RAD17 | RAD17 Entrez, Source | RAD17 homolog (S. pombe) | 6622 | -0.006 | -0.3203 | No |
| 23 | OGG1 | OGG1 Entrez, Source | 8-oxoguanine DNA glycosylase | 7089 | -0.014 | -0.3536 | No |
| 24 | SOD1 | SOD1 Entrez, Source | superoxide dismutase 1, soluble (amyotrophic lateral sclerosis 1 (adult)) | 7093 | -0.014 | -0.3520 | No |
| 25 | MNAT1 | MNAT1 Entrez, Source | menage a trois homolog 1, cyclin H assembly factor (Xenopus laevis) | 7213 | -0.016 | -0.3588 | No |
| 26 | DDB1 | DDB1 Entrez, Source | damage-specific DNA binding protein 1, 127kDa | 7256 | -0.017 | -0.3597 | No |
| 27 | APC | APC Entrez, Source | adenomatosis polyposis coli | 7641 | -0.024 | -0.3854 | No |
| 28 | RAD50 | RAD50 Entrez, Source | RAD50 homolog (S. cerevisiae) | 8016 | -0.032 | -0.4094 | No |
| 29 | HTATIP | HTATIP Entrez, Source | HIV-1 Tat interacting protein, 60kDa | 8212 | -0.035 | -0.4193 | No |
| 30 | ATXN3 | ATXN3 Entrez, Source | ataxin 3 | 8313 | -0.038 | -0.4218 | No |
| 31 | TNP1 | TNP1 Entrez, Source | transition protein 1 (during histone to protamine replacement) | 8326 | -0.038 | -0.4177 | No |
| 32 | CSNK1D | CSNK1D Entrez, Source | casein kinase 1, delta | 8498 | -0.041 | -0.4251 | No |
| 33 | NBN | NBN Entrez, Source | nibrin | 8672 | -0.045 | -0.4321 | No |
| 34 | VCP | VCP Entrez, Source | valosin-containing protein | 9171 | -0.055 | -0.4623 | No |
| 35 | RAD23B | RAD23B Entrez, Source | RAD23 homolog B (S. cerevisiae) | 9175 | -0.055 | -0.4551 | No |
| 36 | FANCC | FANCC Entrez, Source | Fanconi anemia, complementation group C | 9324 | -0.059 | -0.4584 | No |
| 37 | UNG | UNG Entrez, Source | uracil-DNA glycosylase | 9382 | -0.060 | -0.4547 | No |
| 38 | CDKN2D | CDKN2D Entrez, Source | cyclin-dependent kinase inhibitor 2D (p19, inhibits CDK4) | 9497 | -0.063 | -0.4549 | No |
| 39 | MUTYH | MUTYH Entrez, Source | mutY homolog (E. coli) | 9606 | -0.065 | -0.4544 | No |
| 40 | IGHMBP2 | IGHMBP2 Entrez, Source | immunoglobulin mu binding protein 2 | 9982 | -0.076 | -0.4725 | Yes |
| 41 | MPG | MPG Entrez, Source | N-methylpurine-DNA glycosylase | 10047 | -0.078 | -0.4669 | Yes |
| 42 | APEX1 | APEX1 Entrez, Source | APEX nuclease (multifunctional DNA repair enzyme) 1 | 10112 | -0.079 | -0.4612 | Yes |
| 43 | ERCC3 | ERCC3 Entrez, Source | excision repair cross-complementing rodent repair deficiency, complementation group 3 (xeroderma pigmentosum group B complementing) | 10351 | -0.086 | -0.4676 | Yes |
| 44 | POLD1 | POLD1 Entrez, Source | polymerase (DNA directed), delta 1, catalytic subunit 125kDa | 10438 | -0.088 | -0.4623 | Yes |
| 45 | CHEK1 | CHEK1 Entrez, Source | CHK1 checkpoint homolog (S. pombe) | 10530 | -0.091 | -0.4571 | Yes |
| 46 | LIG3 | LIG3 Entrez, Source | ligase III, DNA, ATP-dependent | 10605 | -0.094 | -0.4502 | Yes |
| 47 | WRNIP1 | WRNIP1 Entrez, Source | Werner helicase interacting protein 1 | 10609 | -0.094 | -0.4379 | Yes |
| 48 | SMC1A | SMC1A Entrez, Source | structural maintenance of chromosomes 1A | 10658 | -0.096 | -0.4288 | Yes |
| 49 | GTF2H4 | GTF2H4 Entrez, Source | general transcription factor IIH, polypeptide 4, 52kDa | 10727 | -0.098 | -0.4208 | Yes |
| 50 | BRCA1 | BRCA1 Entrez, Source | breast cancer 1, early onset | 11096 | -0.112 | -0.4337 | Yes |
| 51 | BRCA2 | BRCA2 Entrez, Source | breast cancer 2, early onset | 11106 | -0.112 | -0.4194 | Yes |
| 52 | TP53 | TP53 Entrez, Source | tumor protein p53 (Li-Fraumeni syndrome) | 11266 | -0.119 | -0.4155 | Yes |
| 53 | TDG | TDG Entrez, Source | thymine-DNA glycosylase | 11393 | -0.124 | -0.4084 | Yes |
| 54 | UBE2V2 | UBE2V2 Entrez, Source | ubiquitin-conjugating enzyme E2 variant 2 | 11529 | -0.130 | -0.4012 | Yes |
| 55 | UBE2A | UBE2A Entrez, Source | ubiquitin-conjugating enzyme E2A (RAD6 homolog) | 11635 | -0.135 | -0.3911 | Yes |
| 56 | RUVBL2 | RUVBL2 Entrez, Source | RuvB-like 2 (E. coli) | 11685 | -0.137 | -0.3765 | Yes |
| 57 | CSNK1E | CSNK1E Entrez, Source | casein kinase 1, epsilon | 11711 | -0.139 | -0.3599 | Yes |
| 58 | PNKP | PNKP Entrez, Source | polynucleotide kinase 3'-phosphatase | 12212 | -0.172 | -0.3746 | Yes |
| 59 | XAB2 | XAB2 Entrez, Source | XPA binding protein 2 | 12377 | -0.186 | -0.3622 | Yes |
| 60 | GADD45A | GADD45A Entrez, Source | growth arrest and DNA-damage-inducible, alpha | 12556 | -0.205 | -0.3482 | Yes |
| 61 | RFC3 | RFC3 Entrez, Source | replication factor C (activator 1) 3, 38kDa | 12712 | -0.226 | -0.3298 | Yes |
| 62 | LIG1 | LIG1 Entrez, Source | ligase I, DNA, ATP-dependent | 12917 | -0.270 | -0.3092 | Yes |
| 63 | FEN1 | FEN1 Entrez, Source | flap structure-specific endonuclease 1 | 12971 | -0.283 | -0.2754 | Yes |
| 64 | MCM7 | MCM7 Entrez, Source | MCM7 minichromosome maintenance deficient 7 (S. cerevisiae) | 13101 | -0.328 | -0.2415 | Yes |
| 65 | MRE11A | MRE11A Entrez, Source | MRE11 meiotic recombination 11 homolog A (S. cerevisiae) | 13163 | -0.372 | -0.1966 | Yes |
| 66 | APTX | APTX Entrez, Source | aprataxin | 13232 | -0.441 | -0.1429 | Yes |
| 67 | DDIT3 | DDIT3 Entrez, Source | DNA-damage-inducible transcript 3 | 13285 | -0.545 | -0.0743 | Yes |
| 68 | GADD45G | GADD45G Entrez, Source | growth arrest and DNA-damage-inducible, gamma | 13297 | -0.589 | 0.0034 | Yes |