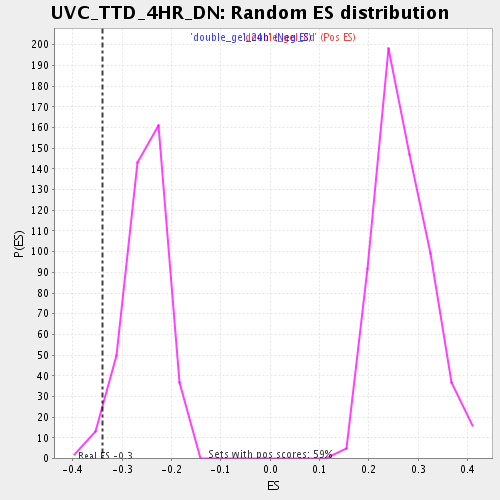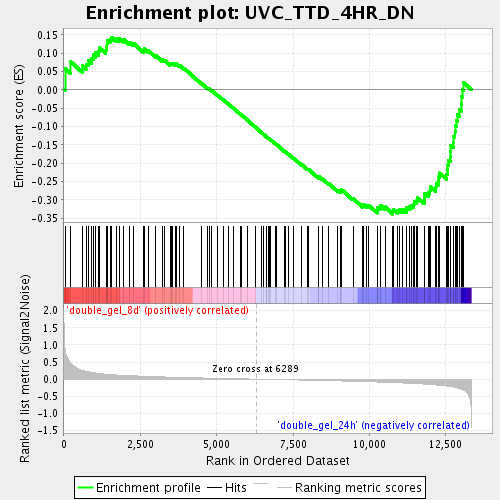
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | double_gel_and_single_gel_rma_expression_values_collapsed_to_symbols.class.cls #double_gel_8d_versus_double_gel_24h.class.cls #double_gel_8d_versus_double_gel_24h_repos |
| Phenotype | class.cls#double_gel_8d_versus_double_gel_24h_repos |
| Upregulated in class | double_gel_24h |
| GeneSet | UVC_TTD_4HR_DN |
| Enrichment Score (ES) | -0.3388554 |
| Normalized Enrichment Score (NES) | -1.3347944 |
| Nominal p-value | 0.024630541 |
| FDR q-value | 0.3738327 |
| FWER p-Value | 1.0 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | CTGF | CTGF Entrez, Source | connective tissue growth factor | 37 | 0.888 | 0.0582 | No |
| 2 | SCHIP1 | SCHIP1 Entrez, Source | schwannomin interacting protein 1 | 217 | 0.472 | 0.0771 | No |
| 3 | PPP2R3A | PPP2R3A Entrez, Source | protein phosphatase 2 (formerly 2A), regulatory subunit B'', alpha | 603 | 0.257 | 0.0657 | No |
| 4 | IGF2BP3 | IGF2BP3 Entrez, Source | insulin-like growth factor 2 mRNA binding protein 3 | 746 | 0.228 | 0.0706 | No |
| 5 | CUTL1 | CUTL1 Entrez, Source | cut-like 1, CCAAT displacement protein (Drosophila) | 807 | 0.212 | 0.0806 | No |
| 6 | TGFB2 | TGFB2 Entrez, Source | transforming growth factor, beta 2 | 916 | 0.193 | 0.0857 | No |
| 7 | F3 | F3 Entrez, Source | coagulation factor III (thromboplastin, tissue factor) | 956 | 0.188 | 0.0957 | No |
| 8 | IL1RAP | IL1RAP Entrez, Source | interleukin 1 receptor accessory protein | 1043 | 0.177 | 0.1013 | No |
| 9 | SIPA1L1 | SIPA1L1 Entrez, Source | signal-induced proliferation-associated 1 like 1 | 1138 | 0.167 | 0.1057 | No |
| 10 | PPAP2B | PPAP2B Entrez, Source | phosphatidic acid phosphatase type 2B | 1157 | 0.164 | 0.1156 | No |
| 11 | MSH3 | MSH3 Entrez, Source | mutS homolog 3 (E. coli) | 1377 | 0.144 | 0.1090 | No |
| 12 | EDG2 | EDG2 Entrez, Source | endothelial differentiation, lysophosphatidic acid G-protein-coupled receptor, 2 | 1399 | 0.143 | 0.1172 | No |
| 13 | VEGFC | VEGFC Entrez, Source | vascular endothelial growth factor C | 1410 | 0.142 | 0.1262 | No |
| 14 | PRKCA | PRKCA Entrez, Source | protein kinase C, alpha | 1423 | 0.141 | 0.1350 | No |
| 15 | BARD1 | BARD1 Entrez, Source | BRCA1 associated RING domain 1 | 1511 | 0.134 | 0.1377 | No |
| 16 | PTPN2 | PTPN2 Entrez, Source | protein tyrosine phosphatase, non-receptor type 2 | 1553 | 0.131 | 0.1436 | No |
| 17 | GNAQ | GNAQ Entrez, Source | guanine nucleotide binding protein (G protein), q polypeptide | 1722 | 0.122 | 0.1393 | No |
| 18 | BMP2K | BMP2K Entrez, Source | BMP2 inducible kinase | 1811 | 0.116 | 0.1406 | No |
| 19 | CNOT4 | CNOT4 Entrez, Source | CCR4-NOT transcription complex, subunit 4 | 1951 | 0.110 | 0.1376 | No |
| 20 | STK39 | STK39 Entrez, Source | serine threonine kinase 39 (STE20/SPS1 homolog, yeast) | 2151 | 0.101 | 0.1295 | No |
| 21 | RGS7 | RGS7 Entrez, Source | regulator of G-protein signalling 7 | 2271 | 0.097 | 0.1272 | No |
| 22 | XRCC4 | XRCC4 Entrez, Source | X-ray repair complementing defective repair in Chinese hamster cells 4 | 2599 | 0.083 | 0.1082 | No |
| 23 | FYN | FYN Entrez, Source | FYN oncogene related to SRC, FGR, YES | 2623 | 0.083 | 0.1121 | No |
| 24 | STK3 | STK3 Entrez, Source | serine/threonine kinase 3 (STE20 homolog, yeast) | 2756 | 0.078 | 0.1075 | No |
| 25 | CEP170 | CEP170 Entrez, Source | centrosomal protein 170kDa | 3004 | 0.071 | 0.0937 | No |
| 26 | FGF2 | FGF2 Entrez, Source | fibroblast growth factor 2 (basic) | 3210 | 0.065 | 0.0827 | No |
| 27 | ID1 | ID1 Entrez, Source | inhibitor of DNA binding 1, dominant negative helix-loop-helix protein | 3286 | 0.063 | 0.0814 | No |
| 28 | IFRD1 | IFRD1 Entrez, Source | interferon-related developmental regulator 1 | 3472 | 0.058 | 0.0714 | No |
| 29 | TLE4 | TLE4 Entrez, Source | transducin-like enhancer of split 4 (E(sp1) homolog, Drosophila) | 3506 | 0.057 | 0.0729 | No |
| 30 | PLEKHC1 | PLEKHC1 Entrez, Source | pleckstrin homology domain containing, family C (with FERM domain) member 1 | 3561 | 0.056 | 0.0726 | No |
| 31 | UBL3 | UBL3 Entrez, Source | ubiquitin-like 3 | 3635 | 0.054 | 0.0708 | No |
| 32 | ZHX3 | ZHX3 Entrez, Source | zinc fingers and homeoboxes 3 | 3677 | 0.053 | 0.0713 | No |
| 33 | SMAD3 | SMAD3 Entrez, Source | SMAD, mothers against DPP homolog 3 (Drosophila) | 3790 | 0.050 | 0.0663 | No |
| 34 | WDR7 | WDR7 Entrez, Source | WD repeat domain 7 | 3924 | 0.047 | 0.0594 | No |
| 35 | RRAS2 | RRAS2 Entrez, Source | related RAS viral (r-ras) oncogene homolog 2 | 4502 | 0.034 | 0.0181 | No |
| 36 | TMEM23 | TMEM23 Entrez, Source | transmembrane protein 23 | 4709 | 0.030 | 0.0046 | No |
| 37 | ORC2L | ORC2L Entrez, Source | origin recognition complex, subunit 2-like (yeast) | 4748 | 0.029 | 0.0037 | No |
| 38 | ATXN1 | ATXN1 Entrez, Source | ataxin 1 | 4845 | 0.027 | -0.0017 | No |
| 39 | PEX14 | PEX14 Entrez, Source | peroxisomal biogenesis factor 14 | 5041 | 0.023 | -0.0148 | No |
| 40 | MYST3 | MYST3 Entrez, Source | MYST histone acetyltransferase (monocytic leukemia) 3 | 5213 | 0.020 | -0.0264 | No |
| 41 | UGCG | UGCG Entrez, Source | UDP-glucose ceramide glucosyltransferase | 5384 | 0.017 | -0.0381 | No |
| 42 | ARNT2 | ARNT2 Entrez, Source | aryl-hydrocarbon receptor nuclear translocator 2 | 5543 | 0.014 | -0.0491 | No |
| 43 | CNOT2 | CNOT2 Entrez, Source | CCR4-NOT transcription complex, subunit 2 | 5786 | 0.009 | -0.0667 | No |
| 44 | KIF2A | KIF2A Entrez, Source | kinesin heavy chain member 2A | 5813 | 0.009 | -0.0681 | No |
| 45 | CTBP2 | CTBP2 Entrez, Source | C-terminal binding protein 2 | 5997 | 0.005 | -0.0816 | No |
| 46 | NUP98 | NUP98 Entrez, Source | nucleoporin 98kDa | 6268 | 0.000 | -0.1020 | No |
| 47 | PTK2 | PTK2 Entrez, Source | PTK2 protein tyrosine kinase 2 | 6465 | -0.003 | -0.1166 | No |
| 48 | AFAP | AFAP Entrez, Source | - | 6545 | -0.005 | -0.1223 | No |
| 49 | SPBC25 | SPBC25 Entrez, Source | spindle pole body component 25 homolog (S. cerevisiae) | 6642 | -0.006 | -0.1291 | No |
| 50 | DLC1 | DLC1 Entrez, Source | deleted in liver cancer 1 | 6645 | -0.006 | -0.1288 | No |
| 51 | EIF4E | EIF4E Entrez, Source | eukaryotic translation initiation factor 4E | 6696 | -0.007 | -0.1321 | No |
| 52 | ACVR1 | ACVR1 Entrez, Source | activin A receptor, type I | 6711 | -0.007 | -0.1327 | No |
| 53 | CBLB | CBLB Entrez, Source | Cas-Br-M (murine) ecotropic retroviral transforming sequence b | 6744 | -0.008 | -0.1345 | No |
| 54 | CASP8 | CASP8 Entrez, Source | caspase 8, apoptosis-related cysteine peptidase | 6929 | -0.011 | -0.1477 | No |
| 55 | SPOP | SPOP Entrez, Source | speckle-type POZ protein | 6946 | -0.011 | -0.1481 | No |
| 56 | MNAT1 | MNAT1 Entrez, Source | menage a trois homolog 1, cyclin H assembly factor (Xenopus laevis) | 7213 | -0.016 | -0.1671 | No |
| 57 | HAS2 | HAS2 Entrez, Source | hyaluronan synthase 2 | 7235 | -0.017 | -0.1676 | No |
| 58 | ZNF148 | ZNF148 Entrez, Source | zinc finger protein 148 | 7364 | -0.019 | -0.1759 | No |
| 59 | GNAI1 | GNAI1 Entrez, Source | guanine nucleotide binding protein (G protein), alpha inhibiting activity polypeptide 1 | 7518 | -0.022 | -0.1860 | No |
| 60 | TSPAN5 | TSPAN5 Entrez, Source | tetraspanin 5 | 7785 | -0.027 | -0.2043 | No |
| 61 | MAP2K4 | MAP2K4 Entrez, Source | mitogen-activated protein kinase kinase 4 | 7969 | -0.031 | -0.2160 | No |
| 62 | PARG | PARG Entrez, Source | poly (ADP-ribose) glycohydrolase | 7997 | -0.031 | -0.2159 | No |
| 63 | MTMR3 | MTMR3 Entrez, Source | myotubularin related protein 3 | 8330 | -0.038 | -0.2384 | No |
| 64 | STRN3 | STRN3 Entrez, Source | striatin, calmodulin binding protein 3 | 8342 | -0.038 | -0.2366 | No |
| 65 | MGAT5 | MGAT5 Entrez, Source | mannosyl (alpha-1,6-)-glycoprotein beta-1,6-N-acetyl-glucosaminyltransferase | 8458 | -0.041 | -0.2425 | No |
| 66 | SYNJ2 | SYNJ2 Entrez, Source | synaptojanin 2 | 8657 | -0.045 | -0.2544 | No |
| 67 | PPP2R2A | PPP2R2A Entrez, Source | protein phosphatase 2 (formerly 2A), regulatory subunit B (PR 52), alpha isoform | 8961 | -0.051 | -0.2739 | No |
| 68 | PKN2 | PKN2 Entrez, Source | protein kinase N2 | 9045 | -0.053 | -0.2765 | No |
| 69 | PTPN12 | PTPN12 Entrez, Source | protein tyrosine phosphatase, non-receptor type 12 | 9058 | -0.053 | -0.2738 | No |
| 70 | FGF5 | FGF5 Entrez, Source | fibroblast growth factor 5 | 9095 | -0.054 | -0.2728 | No |
| 71 | GULP1 | GULP1 Entrez, Source | GULP, engulfment adaptor PTB domain containing 1 | 9461 | -0.062 | -0.2961 | No |
| 72 | ACVR2A | ACVR2A Entrez, Source | activin A receptor, type IIA | 9778 | -0.070 | -0.3152 | No |
| 73 | ATP2B1 | ATP2B1 Entrez, Source | ATPase, Ca++ transporting, plasma membrane 1 | 9817 | -0.071 | -0.3132 | No |
| 74 | SLIT1 | SLIT1 Entrez, Source | slit homolog 1 (Drosophila) | 9915 | -0.074 | -0.3154 | No |
| 75 | NCOA3 | NCOA3 Entrez, Source | nuclear receptor coactivator 3 | 9971 | -0.076 | -0.3144 | No |
| 76 | MTF2 | MTF2 Entrez, Source | metal response element binding transcription factor 2 | 10257 | -0.083 | -0.3302 | No |
| 77 | LYN | LYN Entrez, Source | v-yes-1 Yamaguchi sarcoma viral related oncogene homolog | 10258 | -0.083 | -0.3245 | No |
| 78 | PUM2 | PUM2 Entrez, Source | pumilio homolog 2 (Drosophila) | 10272 | -0.084 | -0.3197 | No |
| 79 | PPP1R12A | PPP1R12A Entrez, Source | protein phosphatase 1, regulatory (inhibitor) subunit 12A | 10350 | -0.086 | -0.3197 | No |
| 80 | MID1 | MID1 Entrez, Source | midline 1 (Opitz/BBB syndrome) | 10356 | -0.086 | -0.3141 | No |
| 81 | MAPK6 | MAPK6 Entrez, Source | mitogen-activated protein kinase 6 | 10508 | -0.090 | -0.3193 | No |
| 82 | PTPRK | PTPRK Entrez, Source | protein tyrosine phosphatase, receptor type, K | 10767 | -0.100 | -0.3320 | Yes |
| 83 | RBM16 | RBM16 Entrez, Source | RNA binding motif protein 16 | 10775 | -0.100 | -0.3257 | Yes |
| 84 | ITSN1 | ITSN1 Entrez, Source | intersectin 1 (SH3 domain protein) | 10914 | -0.105 | -0.3289 | Yes |
| 85 | CLASP1 | CLASP1 Entrez, Source | cytoplasmic linker associated protein 1 | 10986 | -0.108 | -0.3268 | Yes |
| 86 | CREBBP | CREBBP Entrez, Source | CREB binding protein (Rubinstein-Taybi syndrome) | 11078 | -0.111 | -0.3261 | Yes |
| 87 | PCTK2 | PCTK2 Entrez, Source | PCTAIRE protein kinase 2 | 11205 | -0.116 | -0.3276 | Yes |
| 88 | ARHGEF7 | ARHGEF7 Entrez, Source | Rho guanine nucleotide exchange factor (GEF) 7 | 11215 | -0.117 | -0.3202 | Yes |
| 89 | MARCH7 | MARCH7 Entrez, Source | membrane-associated ring finger (C3HC4) 7 | 11303 | -0.121 | -0.3185 | Yes |
| 90 | CEP350 | CEP350 Entrez, Source | centrosomal protein 350kDa | 11387 | -0.124 | -0.3163 | Yes |
| 91 | ROCK2 | ROCK2 Entrez, Source | Rho-associated, coiled-coil containing protein kinase 2 | 11450 | -0.127 | -0.3122 | Yes |
| 92 | NIPBL | NIPBL Entrez, Source | Nipped-B homolog (Drosophila) | 11464 | -0.127 | -0.3045 | Yes |
| 93 | TCF12 | TCF12 Entrez, Source | transcription factor 12 (HTF4, helix-loop-helix transcription factors 4) | 11550 | -0.131 | -0.3019 | Yes |
| 94 | HOMER1 | HOMER1 Entrez, Source | homer homolog 1 (Drosophila) | 11571 | -0.132 | -0.2943 | Yes |
| 95 | CLASP2 | CLASP2 Entrez, Source | cytoplasmic linker associated protein 2 | 11792 | -0.144 | -0.3010 | Yes |
| 96 | PAFAH1B1 | PAFAH1B1 Entrez, Source | platelet-activating factor acetylhydrolase, isoform Ib, alpha subunit 45kDa | 11805 | -0.145 | -0.2920 | Yes |
| 97 | OGT | OGT Entrez, Source | O-linked N-acetylglucosamine (GlcNAc) transferase (UDP-N-acetylglucosamine:polypeptide-N-acetylglucosaminyl transferase) | 11813 | -0.145 | -0.2825 | Yes |
| 98 | RBM39 | RBM39 Entrez, Source | RNA binding motif protein 39 | 11924 | -0.152 | -0.2804 | Yes |
| 99 | NUP153 | NUP153 Entrez, Source | nucleoporin 153kDa | 11979 | -0.156 | -0.2738 | Yes |
| 100 | GSK3B | GSK3B Entrez, Source | glycogen synthase kinase 3 beta | 11989 | -0.156 | -0.2637 | Yes |
| 101 | RSN | RSN Entrez, Source | restin (Reed-Steinberg cell-expressed intermediate filament-associated protein) | 12170 | -0.169 | -0.2657 | Yes |
| 102 | PRIM2A | PRIM2A Entrez, Source | primase, polypeptide 2A, 58kDa | 12178 | -0.170 | -0.2545 | Yes |
| 103 | ABI1 | ABI1 Entrez, Source | abl-interactor 1 | 12248 | -0.175 | -0.2477 | Yes |
| 104 | TUBGCP3 | TUBGCP3 Entrez, Source | tubulin, gamma complex associated protein 3 | 12272 | -0.177 | -0.2373 | Yes |
| 105 | ARIH1 | ARIH1 Entrez, Source | ariadne homolog, ubiquitin-conjugating enzyme E2 binding protein, 1 (Drosophila) | 12297 | -0.179 | -0.2267 | Yes |
| 106 | ZNF292 | ZNF292 Entrez, Source | zinc finger protein 292 | 12529 | -0.202 | -0.2303 | Yes |
| 107 | RUNX1 | RUNX1 Entrez, Source | runt-related transcription factor 1 (acute myeloid leukemia 1; aml1 oncogene) | 12547 | -0.204 | -0.2176 | Yes |
| 108 | PARD3 | PARD3 Entrez, Source | par-3 partitioning defective 3 homolog (C. elegans) | 12567 | -0.207 | -0.2048 | Yes |
| 109 | PRKCBP1 | PRKCBP1 Entrez, Source | protein kinase C binding protein 1 | 12589 | -0.209 | -0.1920 | Yes |
| 110 | ATF2 | ATF2 Entrez, Source | activating transcription factor 2 | 12642 | -0.216 | -0.1811 | Yes |
| 111 | DYRK1A | DYRK1A Entrez, Source | dual-specificity tyrosine-(Y)-phosphorylation regulated kinase 1A | 12656 | -0.218 | -0.1670 | Yes |
| 112 | CRY1 | CRY1 Entrez, Source | cryptochrome 1 (photolyase-like) | 12664 | -0.219 | -0.1525 | Yes |
| 113 | HNRPDL | HNRPDL Entrez, Source | heterogeneous nuclear ribonucleoprotein D-like | 12742 | -0.232 | -0.1424 | Yes |
| 114 | ZRF1 | ZRF1 Entrez, Source | zuotin related factor 1 | 12750 | -0.233 | -0.1269 | Yes |
| 115 | MDN1 | MDN1 Entrez, Source | MDN1, midasin homolog (yeast) | 12799 | -0.241 | -0.1139 | Yes |
| 116 | SLC4A7 | SLC4A7 Entrez, Source | solute carrier family 4, sodium bicarbonate cotransporter, member 7 | 12805 | -0.242 | -0.0976 | Yes |
| 117 | REV3L | REV3L Entrez, Source | REV3-like, catalytic subunit of DNA polymerase zeta (yeast) | 12847 | -0.253 | -0.0833 | Yes |
| 118 | BHLHB2 | BHLHB2 Entrez, Source | basic helix-loop-helix domain containing, class B, 2 | 12874 | -0.260 | -0.0674 | Yes |
| 119 | EGFR | EGFR Entrez, Source | epidermal growth factor receptor (erythroblastic leukemia viral (v-erb-b) oncogene homolog, avian) | 12946 | -0.276 | -0.0538 | Yes |
| 120 | FUBP1 | FUBP1 Entrez, Source | far upstream element (FUSE) binding protein 1 | 12999 | -0.292 | -0.0377 | Yes |
| 121 | TRIO | TRIO Entrez, Source | triple functional domain (PTPRF interacting) | 13021 | -0.298 | -0.0188 | Yes |
| 122 | PHLPP | PHLPP Entrez, Source | PH domain and leucine rich repeat protein phosphatase | 13038 | -0.305 | 0.0010 | Yes |
| 123 | KLF10 | KLF10 Entrez, Source | Kruppel-like factor 10 | 13083 | -0.319 | 0.0196 | Yes |