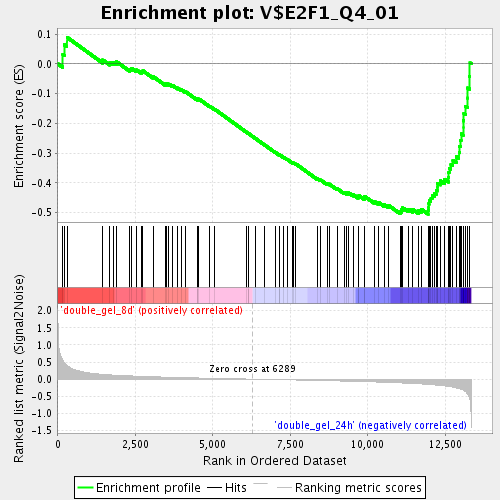
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | double_gel_and_single_gel_rma_expression_values_collapsed_to_symbols.class.cls #double_gel_8d_versus_double_gel_24h.class.cls #double_gel_8d_versus_double_gel_24h_repos |
| Phenotype | class.cls#double_gel_8d_versus_double_gel_24h_repos |
| Upregulated in class | double_gel_24h |
| GeneSet | V$E2F1_Q4_01 |
| Enrichment Score (ES) | -0.5046724 |
| Normalized Enrichment Score (NES) | -1.8792253 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.024330975 |
| FWER p-Value | 0.526 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | DPYSL2 | DPYSL2 Entrez, Source | dihydropyrimidinase-like 2 | 158 | 0.552 | 0.0308 | No |
| 2 | USP2 | USP2 Entrez, Source | ubiquitin specific peptidase 2 | 207 | 0.480 | 0.0643 | No |
| 3 | PAQR4 | PAQR4 Entrez, Source | progestin and adipoQ receptor family member IV | 293 | 0.407 | 0.0894 | No |
| 4 | YWHAQ | YWHAQ Entrez, Source | tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, theta polypeptide | 1447 | 0.139 | 0.0132 | No |
| 5 | SLC38A1 | SLC38A1 Entrez, Source | solute carrier family 38, member 1 | 1676 | 0.124 | 0.0056 | No |
| 6 | DAXX | DAXX Entrez, Source | death-associated protein 6 | 1796 | 0.117 | 0.0057 | No |
| 7 | ITGA1 | ITGA1 Entrez, Source | integrin, alpha 1 | 1891 | 0.112 | 0.0073 | No |
| 8 | PCSK1 | PCSK1 Entrez, Source | proprotein convertase subtilisin/kexin type 1 | 2319 | 0.095 | -0.0176 | No |
| 9 | KIAA0101 | KIAA0101 Entrez, Source | KIAA0101 | 2376 | 0.093 | -0.0147 | No |
| 10 | FBXO9 | FBXO9 Entrez, Source | F-box protein 9 | 2539 | 0.086 | -0.0203 | No |
| 11 | ING3 | ING3 Entrez, Source | inhibitor of growth family, member 3 | 2696 | 0.080 | -0.0258 | No |
| 12 | PIK3R3 | PIK3R3 Entrez, Source | phosphoinositide-3-kinase, regulatory subunit 3 (p55, gamma) | 2744 | 0.079 | -0.0233 | No |
| 13 | RAB11B | RAB11B Entrez, Source | RAB11B, member RAS oncogene family | 3071 | 0.069 | -0.0425 | No |
| 14 | ID3 | ID3 Entrez, Source | inhibitor of DNA binding 3, dominant negative helix-loop-helix protein | 3479 | 0.058 | -0.0687 | No |
| 15 | TLE4 | TLE4 Entrez, Source | transducin-like enhancer of split 4 (E(sp1) homolog, Drosophila) | 3506 | 0.057 | -0.0662 | No |
| 16 | KCNA6 | KCNA6 Entrez, Source | potassium voltage-gated channel, shaker-related subfamily, member 6 | 3580 | 0.055 | -0.0675 | No |
| 17 | EGR3 | EGR3 Entrez, Source | early growth response 3 | 3704 | 0.052 | -0.0727 | No |
| 18 | PPP1R9B | PPP1R9B Entrez, Source | protein phosphatase 1, regulatory subunit 9B, spinophilin | 3862 | 0.048 | -0.0808 | No |
| 19 | DMRT1 | DMRT1 Entrez, Source | doublesex and mab-3 related transcription factor 1 | 3981 | 0.045 | -0.0862 | No |
| 20 | CDC25A | CDC25A Entrez, Source | cell division cycle 25A | 4102 | 0.042 | -0.0920 | No |
| 21 | KLF5 | KLF5 Entrez, Source | Kruppel-like factor 5 (intestinal) | 4503 | 0.034 | -0.1196 | No |
| 22 | SLC6A4 | SLC6A4 Entrez, Source | solute carrier family 6 (neurotransmitter transporter, serotonin), member 4 | 4511 | 0.034 | -0.1175 | No |
| 23 | ATRX | ATRX Entrez, Source | alpha thalassemia/mental retardation syndrome X-linked (RAD54 homolog, S. cerevisiae) | 4548 | 0.033 | -0.1177 | No |
| 24 | CHGB | CHGB Entrez, Source | chromogranin B (secretogranin 1) | 4906 | 0.026 | -0.1426 | No |
| 25 | PVRL1 | PVRL1 Entrez, Source | poliovirus receptor-related 1 (herpesvirus entry mediator C; nectin) | 5060 | 0.023 | -0.1524 | No |
| 26 | POLA2 | POLA2 Entrez, Source | polymerase (DNA directed), alpha 2 (70kD subunit) | 6089 | 0.003 | -0.2297 | No |
| 27 | ACBD6 | ACBD6 Entrez, Source | acyl-Coenzyme A binding domain containing 6 | 6149 | 0.002 | -0.2340 | No |
| 28 | GATA3 | GATA3 Entrez, Source | GATA binding protein 3 | 6151 | 0.002 | -0.2339 | No |
| 29 | H2AFZ | H2AFZ Entrez, Source | H2A histone family, member Z | 6380 | -0.001 | -0.2510 | No |
| 30 | NEGR1 | NEGR1 Entrez, Source | neuronal growth regulator 1 | 6659 | -0.007 | -0.2715 | No |
| 31 | TOP1 | TOP1 Entrez, Source | topoisomerase (DNA) I | 7017 | -0.013 | -0.2974 | No |
| 32 | KBTBD7 | KBTBD7 Entrez, Source | kelch repeat and BTB (POZ) domain containing 7 | 7148 | -0.015 | -0.3061 | No |
| 33 | UXT | UXT Entrez, Source | ubiquitously-expressed transcript | 7154 | -0.015 | -0.3053 | No |
| 34 | USP52 | USP52 Entrez, Source | ubiquitin specific peptidase 52 | 7291 | -0.018 | -0.3142 | No |
| 35 | FBXL20 | FBXL20 Entrez, Source | F-box and leucine-rich repeat protein 20 | 7404 | -0.020 | -0.3211 | No |
| 36 | KCND2 | KCND2 Entrez, Source | potassium voltage-gated channel, Shal-related subfamily, member 2 | 7584 | -0.023 | -0.3328 | No |
| 37 | E2F1 | E2F1 Entrez, Source | E2F transcription factor 1 | 7606 | -0.024 | -0.3326 | No |
| 38 | NDUFA11 | NDUFA11 Entrez, Source | NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 11, 14.7kDa | 7683 | -0.025 | -0.3364 | No |
| 39 | CTCF | CTCF Entrez, Source | CCCTC-binding factor (zinc finger protein) | 8374 | -0.039 | -0.3855 | No |
| 40 | DNMT1 | DNMT1 Entrez, Source | DNA (cytosine-5-)-methyltransferase 1 | 8474 | -0.041 | -0.3898 | No |
| 41 | HOXC10 | HOXC10 Entrez, Source | homeobox C10 | 8695 | -0.045 | -0.4029 | No |
| 42 | NRP2 | NRP2 Entrez, Source | neuropilin 2 | 8752 | -0.047 | -0.4035 | No |
| 43 | CDCA7 | CDCA7 Entrez, Source | cell division cycle associated 7 | 9009 | -0.052 | -0.4188 | No |
| 44 | FMO4 | FMO4 Entrez, Source | flavin containing monooxygenase 4 | 9249 | -0.057 | -0.4324 | No |
| 45 | FANCC | FANCC Entrez, Source | Fanconi anemia, complementation group C | 9324 | -0.059 | -0.4334 | No |
| 46 | UNG | UNG Entrez, Source | uracil-DNA glycosylase | 9382 | -0.060 | -0.4331 | No |
| 47 | RALY | RALY Entrez, Source | RNA binding protein, autoantigenic (hnRNP-associated with lethal yellow homolog (mouse)) | 9531 | -0.064 | -0.4393 | No |
| 48 | EPHB1 | EPHB1 Entrez, Source | EPH receptor B1 | 9688 | -0.068 | -0.4459 | No |
| 49 | RFC1 | RFC1 Entrez, Source | replication factor C (activator 1) 1, 145kDa | 9713 | -0.068 | -0.4424 | No |
| 50 | SMAD6 | SMAD6 Entrez, Source | SMAD, mothers against DPP homolog 6 (Drosophila) | 9887 | -0.073 | -0.4498 | No |
| 51 | DMD | DMD Entrez, Source | dystrophin (muscular dystrophy, Duchenne and Becker types) | 9896 | -0.074 | -0.4447 | No |
| 52 | MXD3 | MXD3 Entrez, Source | MAX dimerization protein 3 | 10205 | -0.082 | -0.4616 | No |
| 53 | HMGN2 | HMGN2 Entrez, Source | high-mobility group nucleosomal binding domain 2 | 10359 | -0.086 | -0.4665 | No |
| 54 | NCOA6 | NCOA6 Entrez, Source | nuclear receptor coactivator 6 | 10541 | -0.091 | -0.4730 | No |
| 55 | NUMA1 | NUMA1 Entrez, Source | nuclear mitotic apparatus protein 1 | 10675 | -0.096 | -0.4756 | No |
| 56 | DDX17 | DDX17 Entrez, Source | DEAD (Asp-Glu-Ala-Asp) box polypeptide 17 | 11050 | -0.110 | -0.4953 | Yes |
| 57 | NR3C2 | NR3C2 Entrez, Source | nuclear receptor subfamily 3, group C, member 2 | 11091 | -0.112 | -0.4897 | Yes |
| 58 | RHD | RHD Entrez, Source | Rh blood group, D antigen | 11128 | -0.113 | -0.4837 | Yes |
| 59 | RPS19 | RPS19 Entrez, Source | ribosomal protein S19 | 11310 | -0.121 | -0.4880 | Yes |
| 60 | PCNA | PCNA Entrez, Source | proliferating cell nuclear antigen | 11449 | -0.127 | -0.4885 | Yes |
| 61 | BMP7 | BMP7 Entrez, Source | bone morphogenetic protein 7 (osteogenic protein 1) | 11644 | -0.136 | -0.4927 | Yes |
| 62 | GLRA3 | GLRA3 Entrez, Source | glycine receptor, alpha 3 | 11725 | -0.140 | -0.4879 | Yes |
| 63 | WEE1 | WEE1 Entrez, Source | WEE1 homolog (S. pombe) | 11948 | -0.154 | -0.4928 | Yes |
| 64 | UFD1L | UFD1L Entrez, Source | ubiquitin fusion degradation 1 like (yeast) | 11964 | -0.154 | -0.4820 | Yes |
| 65 | HTF9C | HTF9C Entrez, Source | - | 11970 | -0.155 | -0.4703 | Yes |
| 66 | NUP153 | NUP153 Entrez, Source | nucleoporin 153kDa | 11979 | -0.156 | -0.4589 | Yes |
| 67 | SFRS2 | SFRS2 Entrez, Source | splicing factor, arginine/serine-rich 2 | 12039 | -0.160 | -0.4509 | Yes |
| 68 | XTP3TPA | XTP3TPA Entrez, Source | - | 12079 | -0.163 | -0.4413 | Yes |
| 69 | SFRS12 | SFRS12 Entrez, Source | splicing factor, arginine/serine-rich 12 | 12159 | -0.169 | -0.4342 | Yes |
| 70 | UCHL1 | UCHL1 Entrez, Source | ubiquitin carboxyl-terminal esterase L1 (ubiquitin thiolesterase) | 12209 | -0.172 | -0.4245 | Yes |
| 71 | STMN1 | STMN1 Entrez, Source | stathmin 1/oncoprotein 18 | 12244 | -0.175 | -0.4136 | Yes |
| 72 | MYC | MYC Entrez, Source | v-myc myelocytomatosis viral oncogene homolog (avian) | 12249 | -0.175 | -0.4003 | Yes |
| 73 | PRPS1 | PRPS1 Entrez, Source | phosphoribosyl pyrophosphate synthetase 1 | 12339 | -0.183 | -0.3929 | Yes |
| 74 | KPNB1 | KPNB1 Entrez, Source | karyopherin (importin) beta 1 | 12477 | -0.196 | -0.3880 | Yes |
| 75 | DHX40 | DHX40 Entrez, Source | DEAH (Asp-Glu-Ala-His) box polypeptide 40 | 12600 | -0.211 | -0.3809 | Yes |
| 76 | IMPDH2 | IMPDH2 Entrez, Source | IMP (inosine monophosphate) dehydrogenase 2 | 12613 | -0.213 | -0.3653 | Yes |
| 77 | USP1 | USP1 Entrez, Source | ubiquitin specific peptidase 1 | 12629 | -0.215 | -0.3498 | Yes |
| 78 | ATP5G2 | ATP5G2 Entrez, Source | ATP synthase, H+ transporting, mitochondrial F0 complex, subunit C2 (subunit 9) | 12683 | -0.222 | -0.3367 | Yes |
| 79 | HMGA1 | HMGA1 Entrez, Source | high mobility group AT-hook 1 | 12748 | -0.232 | -0.3235 | Yes |
| 80 | NCL | NCL Entrez, Source | nucleolin | 12857 | -0.256 | -0.3119 | Yes |
| 81 | PTMA | PTMA Entrez, Source | prothymosin, alpha (gene sequence 28) | 12945 | -0.275 | -0.2971 | Yes |
| 82 | NUP155 | NUP155 Entrez, Source | nucleoporin 155kDa | 12960 | -0.279 | -0.2766 | Yes |
| 83 | NASP | NASP Entrez, Source | nuclear autoantigenic sperm protein (histone-binding) | 12994 | -0.291 | -0.2565 | Yes |
| 84 | PIM1 | PIM1 Entrez, Source | pim-1 oncogene | 13014 | -0.296 | -0.2350 | Yes |
| 85 | HNRPA1 | HNRPA1 Entrez, Source | heterogeneous nuclear ribonucleoprotein A1 | 13080 | -0.318 | -0.2153 | Yes |
| 86 | PELO | PELO Entrez, Source | pelota homolog (Drosophila) | 13082 | -0.319 | -0.1907 | Yes |
| 87 | MCM7 | MCM7 Entrez, Source | MCM7 minichromosome maintenance deficient 7 (S. cerevisiae) | 13101 | -0.328 | -0.1667 | Yes |
| 88 | CASP2 | CASP2 Entrez, Source | caspase 2, apoptosis-related cysteine peptidase (neural precursor cell expressed, developmentally down-regulated 2) | 13151 | -0.360 | -0.1425 | Yes |
| 89 | DCK | DCK Entrez, Source | deoxycytidine kinase | 13222 | -0.428 | -0.1146 | Yes |
| 90 | HNRPD | HNRPD Entrez, Source | heterogeneous nuclear ribonucleoprotein D (AU-rich element RNA binding protein 1, 37kDa) | 13227 | -0.440 | -0.0809 | Yes |
| 91 | MCM6 | MCM6 Entrez, Source | MCM6 minichromosome maintenance deficient 6 (MIS5 homolog, S. pombe) (S. cerevisiae) | 13288 | -0.553 | -0.0426 | Yes |
| 92 | MAT2A | MAT2A Entrez, Source | methionine adenosyltransferase II, alpha | 13299 | -0.601 | 0.0032 | Yes |