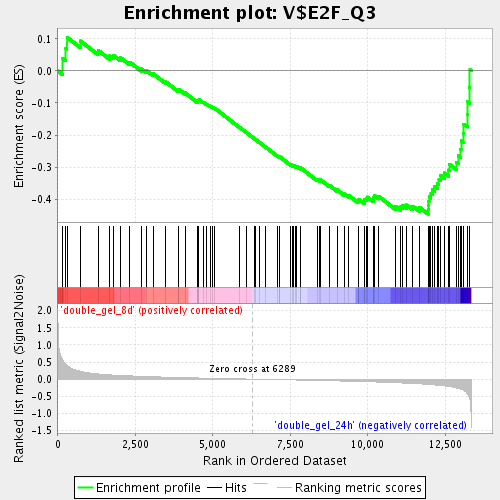
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | double_gel_and_single_gel_rma_expression_values_collapsed_to_symbols.class.cls #double_gel_8d_versus_double_gel_24h.class.cls #double_gel_8d_versus_double_gel_24h_repos |
| Phenotype | class.cls#double_gel_8d_versus_double_gel_24h_repos |
| Upregulated in class | double_gel_24h |
| GeneSet | V$E2F_Q3 |
| Enrichment Score (ES) | -0.44622245 |
| Normalized Enrichment Score (NES) | -1.682609 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.109304726 |
| FWER p-Value | 0.999 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | DPYSL2 | DPYSL2 Entrez, Source | dihydropyrimidinase-like 2 | 158 | 0.552 | 0.0382 | No |
| 2 | KLF4 | KLF4 Entrez, Source | Kruppel-like factor 4 (gut) | 255 | 0.437 | 0.0706 | No |
| 3 | PAQR4 | PAQR4 Entrez, Source | progestin and adipoQ receptor family member IV | 293 | 0.407 | 0.1049 | No |
| 4 | FLI1 | FLI1 Entrez, Source | Friend leukemia virus integration 1 | 724 | 0.231 | 0.0934 | No |
| 5 | ONECUT1 | ONECUT1 Entrez, Source | one cut domain, family member 1 | 1295 | 0.151 | 0.0641 | No |
| 6 | SLC38A1 | SLC38A1 Entrez, Source | solute carrier family 38, member 1 | 1676 | 0.124 | 0.0467 | No |
| 7 | SEZ6 | SEZ6 Entrez, Source | seizure related 6 homolog (mouse) | 1784 | 0.118 | 0.0493 | No |
| 8 | ASCL1 | ASCL1 Entrez, Source | achaete-scute complex-like 1 (Drosophila) | 2026 | 0.107 | 0.0408 | No |
| 9 | PCSK1 | PCSK1 Entrez, Source | proprotein convertase subtilisin/kexin type 1 | 2319 | 0.095 | 0.0274 | No |
| 10 | ING3 | ING3 Entrez, Source | inhibitor of growth family, member 3 | 2696 | 0.080 | 0.0063 | No |
| 11 | ORC1L | ORC1L Entrez, Source | origin recognition complex, subunit 1-like (yeast) | 2867 | 0.075 | 0.0003 | No |
| 12 | RAB11B | RAB11B Entrez, Source | RAB11B, member RAS oncogene family | 3071 | 0.069 | -0.0087 | No |
| 13 | ID3 | ID3 Entrez, Source | inhibitor of DNA binding 3, dominant negative helix-loop-helix protein | 3479 | 0.058 | -0.0341 | No |
| 14 | LHX5 | LHX5 Entrez, Source | LIM homeobox 5 | 3894 | 0.047 | -0.0610 | No |
| 15 | DLST | DLST Entrez, Source | dihydrolipoamide S-succinyltransferase (E2 component of 2-oxo-glutarate complex) | 3905 | 0.047 | -0.0575 | No |
| 16 | CDC25A | CDC25A Entrez, Source | cell division cycle 25A | 4102 | 0.042 | -0.0685 | No |
| 17 | KLF5 | KLF5 Entrez, Source | Kruppel-like factor 5 (intestinal) | 4503 | 0.034 | -0.0956 | No |
| 18 | SLC6A4 | SLC6A4 Entrez, Source | solute carrier family 6 (neurotransmitter transporter, serotonin), member 4 | 4511 | 0.034 | -0.0931 | No |
| 19 | ATRX | ATRX Entrez, Source | alpha thalassemia/mental retardation syndrome X-linked (RAD54 homolog, S. cerevisiae) | 4548 | 0.033 | -0.0928 | No |
| 20 | PCSK4 | PCSK4 Entrez, Source | proprotein convertase subtilisin/kexin type 4 | 4552 | 0.033 | -0.0900 | No |
| 21 | BARHL1 | BARHL1 Entrez, Source | BarH-like 1 (Drosophila) | 4685 | 0.030 | -0.0972 | No |
| 22 | SLC25A3 | SLC25A3 Entrez, Source | solute carrier family 25 (mitochondrial carrier; phosphate carrier), member 3 | 4806 | 0.028 | -0.1037 | No |
| 23 | SYT11 | SYT11 Entrez, Source | synaptotagmin XI | 4918 | 0.026 | -0.1098 | No |
| 24 | PPP1CC | PPP1CC Entrez, Source | protein phosphatase 1, catalytic subunit, gamma isoform | 4996 | 0.024 | -0.1134 | No |
| 25 | PVRL1 | PVRL1 Entrez, Source | poliovirus receptor-related 1 (herpesvirus entry mediator C; nectin) | 5060 | 0.023 | -0.1161 | No |
| 26 | TBX3 | TBX3 Entrez, Source | T-box 3 (ulnar mammary syndrome) | 5869 | 0.008 | -0.1763 | No |
| 27 | POLA2 | POLA2 Entrez, Source | polymerase (DNA directed), alpha 2 (70kD subunit) | 6089 | 0.003 | -0.1925 | No |
| 28 | LASP1 | LASP1 Entrez, Source | LIM and SH3 protein 1 | 6351 | -0.001 | -0.2121 | No |
| 29 | H2AFZ | H2AFZ Entrez, Source | H2A histone family, member Z | 6380 | -0.001 | -0.2141 | No |
| 30 | BAT3 | BAT3 Entrez, Source | HLA-B associated transcript 3 | 6518 | -0.004 | -0.2241 | No |
| 31 | CTDSP1 | CTDSP1 Entrez, Source | CTD (carboxy-terminal domain, RNA polymerase II, polypeptide A) small phosphatase 1 | 6700 | -0.007 | -0.2371 | No |
| 32 | GRIK2 | GRIK2 Entrez, Source | glutamate receptor, ionotropic, kainate 2 | 7092 | -0.014 | -0.2653 | No |
| 33 | KBTBD7 | KBTBD7 Entrez, Source | kelch repeat and BTB (POZ) domain containing 7 | 7148 | -0.015 | -0.2681 | No |
| 34 | UXT | UXT Entrez, Source | ubiquitously-expressed transcript | 7154 | -0.015 | -0.2672 | No |
| 35 | CACNA1G | CACNA1G Entrez, Source | calcium channel, voltage-dependent, alpha 1G subunit | 7509 | -0.022 | -0.2919 | No |
| 36 | KCND2 | KCND2 Entrez, Source | potassium voltage-gated channel, Shal-related subfamily, member 2 | 7584 | -0.023 | -0.2954 | No |
| 37 | E2F1 | E2F1 Entrez, Source | E2F transcription factor 1 | 7606 | -0.024 | -0.2948 | No |
| 38 | NDUFA11 | NDUFA11 Entrez, Source | NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 11, 14.7kDa | 7683 | -0.025 | -0.2983 | No |
| 39 | NPR3 | NPR3 Entrez, Source | natriuretic peptide receptor C/guanylate cyclase C (atrionatriuretic peptide receptor C) | 7707 | -0.025 | -0.2977 | No |
| 40 | KCNA1 | KCNA1 Entrez, Source | potassium voltage-gated channel, shaker-related subfamily, member 1 (episodic ataxia with myokymia) | 7818 | -0.028 | -0.3035 | No |
| 41 | SHMT1 | SHMT1 Entrez, Source | serine hydroxymethyltransferase 1 (soluble) | 7827 | -0.028 | -0.3016 | No |
| 42 | CTCF | CTCF Entrez, Source | CCCTC-binding factor (zinc finger protein) | 8374 | -0.039 | -0.3392 | No |
| 43 | PELP1 | PELP1 Entrez, Source | proline, glutamic acid and leucine rich protein 1 | 8455 | -0.041 | -0.3416 | No |
| 44 | DNMT1 | DNMT1 Entrez, Source | DNA (cytosine-5-)-methyltransferase 1 | 8474 | -0.041 | -0.3392 | No |
| 45 | NRP2 | NRP2 Entrez, Source | neuropilin 2 | 8752 | -0.047 | -0.3559 | No |
| 46 | CDCA7 | CDCA7 Entrez, Source | cell division cycle associated 7 | 9009 | -0.052 | -0.3704 | No |
| 47 | FMO4 | FMO4 Entrez, Source | flavin containing monooxygenase 4 | 9249 | -0.057 | -0.3833 | No |
| 48 | UNG | UNG Entrez, Source | uracil-DNA glycosylase | 9382 | -0.060 | -0.3878 | No |
| 49 | EPHB1 | EPHB1 Entrez, Source | EPH receptor B1 | 9688 | -0.068 | -0.4047 | No |
| 50 | RFC1 | RFC1 Entrez, Source | replication factor C (activator 1) 1, 145kDa | 9713 | -0.068 | -0.4003 | No |
| 51 | SMAD6 | SMAD6 Entrez, Source | SMAD, mothers against DPP homolog 6 (Drosophila) | 9887 | -0.073 | -0.4067 | No |
| 52 | DMD | DMD Entrez, Source | dystrophin (muscular dystrophy, Duchenne and Becker types) | 9896 | -0.074 | -0.4006 | No |
| 53 | ADCY8 | ADCY8 Entrez, Source | adenylate cyclase 8 (brain) | 9950 | -0.075 | -0.3978 | No |
| 54 | PHF12 | PHF12 Entrez, Source | PHD finger protein 12 | 9978 | -0.076 | -0.3929 | No |
| 55 | CLTA | CLTA Entrez, Source | clathrin, light chain (Lca) | 10175 | -0.081 | -0.4003 | No |
| 56 | RBBP7 | RBBP7 Entrez, Source | retinoblastoma binding protein 7 | 10181 | -0.081 | -0.3933 | No |
| 57 | MXD3 | MXD3 Entrez, Source | MAX dimerization protein 3 | 10205 | -0.082 | -0.3877 | No |
| 58 | HMGN2 | HMGN2 Entrez, Source | high-mobility group nucleosomal binding domain 2 | 10359 | -0.086 | -0.3914 | No |
| 59 | TCP1 | TCP1 Entrez, Source | t-complex 1 | 10898 | -0.105 | -0.4224 | No |
| 60 | DDX17 | DDX17 Entrez, Source | DEAD (Asp-Glu-Ala-Asp) box polypeptide 17 | 11050 | -0.110 | -0.4238 | No |
| 61 | RHD | RHD Entrez, Source | Rh blood group, D antigen | 11128 | -0.113 | -0.4194 | No |
| 62 | TFRC | TFRC Entrez, Source | transferrin receptor (p90, CD71) | 11249 | -0.118 | -0.4177 | No |
| 63 | PCNA | PCNA Entrez, Source | proliferating cell nuclear antigen | 11449 | -0.127 | -0.4212 | No |
| 64 | SSBP3 | SSBP3 Entrez, Source | single stranded DNA binding protein 3 | 11675 | -0.137 | -0.4257 | No |
| 65 | WEE1 | WEE1 Entrez, Source | WEE1 homolog (S. pombe) | 11948 | -0.154 | -0.4323 | Yes |
| 66 | UFD1L | UFD1L Entrez, Source | ubiquitin fusion degradation 1 like (yeast) | 11964 | -0.154 | -0.4194 | Yes |
| 67 | HTF9C | HTF9C Entrez, Source | - | 11970 | -0.155 | -0.4057 | Yes |
| 68 | NUP153 | NUP153 Entrez, Source | nucleoporin 153kDa | 11979 | -0.156 | -0.3921 | Yes |
| 69 | SFRS2 | SFRS2 Entrez, Source | splicing factor, arginine/serine-rich 2 | 12039 | -0.160 | -0.3820 | Yes |
| 70 | XTP3TPA | XTP3TPA Entrez, Source | - | 12079 | -0.163 | -0.3702 | Yes |
| 71 | SFRS12 | SFRS12 Entrez, Source | splicing factor, arginine/serine-rich 12 | 12159 | -0.169 | -0.3608 | Yes |
| 72 | STMN1 | STMN1 Entrez, Source | stathmin 1/oncoprotein 18 | 12244 | -0.175 | -0.3513 | Yes |
| 73 | GCH1 | GCH1 Entrez, Source | GTP cyclohydrolase 1 (dopa-responsive dystonia) | 12298 | -0.179 | -0.3390 | Yes |
| 74 | PRPS1 | PRPS1 Entrez, Source | phosphoribosyl pyrophosphate synthetase 1 | 12339 | -0.183 | -0.3254 | Yes |
| 75 | KPNB1 | KPNB1 Entrez, Source | karyopherin (importin) beta 1 | 12477 | -0.196 | -0.3179 | Yes |
| 76 | IMPDH2 | IMPDH2 Entrez, Source | IMP (inosine monophosphate) dehydrogenase 2 | 12613 | -0.213 | -0.3088 | Yes |
| 77 | USP1 | USP1 Entrez, Source | ubiquitin specific peptidase 1 | 12629 | -0.215 | -0.2904 | Yes |
| 78 | NCL | NCL Entrez, Source | nucleolin | 12857 | -0.256 | -0.2843 | Yes |
| 79 | LIG1 | LIG1 Entrez, Source | ligase I, DNA, ATP-dependent | 12917 | -0.270 | -0.2642 | Yes |
| 80 | NASP | NASP Entrez, Source | nuclear autoantigenic sperm protein (histone-binding) | 12994 | -0.291 | -0.2435 | Yes |
| 81 | PIM1 | PIM1 Entrez, Source | pim-1 oncogene | 13014 | -0.296 | -0.2180 | Yes |
| 82 | HNRPA1 | HNRPA1 Entrez, Source | heterogeneous nuclear ribonucleoprotein A1 | 13080 | -0.318 | -0.1941 | Yes |
| 83 | MCM7 | MCM7 Entrez, Source | MCM7 minichromosome maintenance deficient 7 (S. cerevisiae) | 13101 | -0.328 | -0.1658 | Yes |
| 84 | DCK | DCK Entrez, Source | deoxycytidine kinase | 13222 | -0.428 | -0.1360 | Yes |
| 85 | HNRPD | HNRPD Entrez, Source | heterogeneous nuclear ribonucleoprotein D (AU-rich element RNA binding protein 1, 37kDa) | 13227 | -0.440 | -0.0964 | Yes |
| 86 | MCM6 | MCM6 Entrez, Source | MCM6 minichromosome maintenance deficient 6 (MIS5 homolog, S. pombe) (S. cerevisiae) | 13288 | -0.553 | -0.0506 | Yes |
| 87 | MAT2A | MAT2A Entrez, Source | methionine adenosyltransferase II, alpha | 13299 | -0.601 | 0.0032 | Yes |