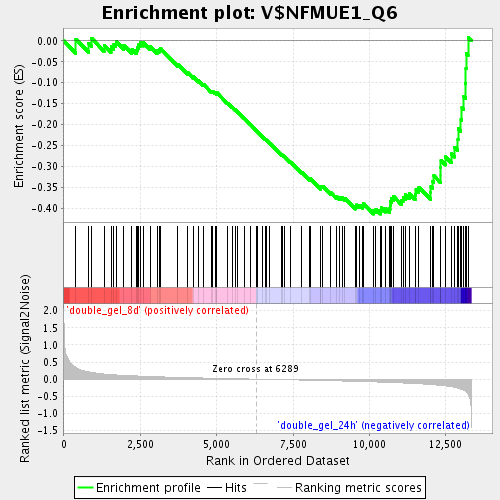
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | double_gel_and_single_gel_rma_expression_values_collapsed_to_symbols.class.cls #double_gel_8d_versus_double_gel_24h.class.cls #double_gel_8d_versus_double_gel_24h_repos |
| Phenotype | class.cls#double_gel_8d_versus_double_gel_24h_repos |
| Upregulated in class | double_gel_24h |
| GeneSet | V$NFMUE1_Q6 |
| Enrichment Score (ES) | -0.41394326 |
| Normalized Enrichment Score (NES) | -1.5742733 |
| Nominal p-value | 0.0051150895 |
| FDR q-value | 0.18326057 |
| FWER p-Value | 1.0 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | CCND1 | CCND1 Entrez, Source | cyclin D1 | 391 | 0.341 | 0.0043 | No |
| 2 | NCDN | NCDN Entrez, Source | neurochondrin | 809 | 0.212 | -0.0062 | No |
| 3 | BCLAF1 | BCLAF1 Entrez, Source | BCL2-associated transcription factor 1 | 898 | 0.196 | 0.0065 | No |
| 4 | CD4 | CD4 Entrez, Source | CD4 molecule | 1315 | 0.149 | -0.0101 | No |
| 5 | ZFP37 | ZFP37 Entrez, Source | zinc finger protein 37 homolog (mouse) | 1543 | 0.132 | -0.0141 | No |
| 6 | FKRP | FKRP Entrez, Source | fukutin related protein | 1635 | 0.126 | -0.0085 | No |
| 7 | DEAF1 | DEAF1 Entrez, Source | deformed epidermal autoregulatory factor 1 (Drosophila) | 1704 | 0.122 | -0.0015 | No |
| 8 | DZIP1 | DZIP1 Entrez, Source | DAZ interacting protein 1 | 1960 | 0.109 | -0.0099 | No |
| 9 | FASTK | FASTK Entrez, Source | Fas-activated serine/threonine kinase | 2227 | 0.098 | -0.0203 | No |
| 10 | APBB1 | APBB1 Entrez, Source | amyloid beta (A4) precursor protein-binding, family B, member 1 (Fe65) | 2372 | 0.093 | -0.0220 | No |
| 11 | HOXA2 | HOXA2 Entrez, Source | homeobox A2 | 2403 | 0.092 | -0.0152 | No |
| 12 | TIA1 | TIA1 Entrez, Source | TIA1 cytotoxic granule-associated RNA binding protein | 2449 | 0.090 | -0.0097 | No |
| 13 | UQCRH | UQCRH Entrez, Source | ubiquinol-cytochrome c reductase hinge protein | 2494 | 0.088 | -0.0043 | No |
| 14 | DGCR2 | DGCR2 Entrez, Source | DiGeorge syndrome critical region gene 2 | 2602 | 0.083 | -0.0041 | No |
| 15 | OTX2 | OTX2 Entrez, Source | orthodenticle homolog 2 (Drosophila) | 2831 | 0.076 | -0.0137 | No |
| 16 | CCNE1 | CCNE1 Entrez, Source | cyclin E1 | 3049 | 0.070 | -0.0232 | No |
| 17 | MARK3 | MARK3 Entrez, Source | MAP/microtubule affinity-regulating kinase 3 | 3113 | 0.068 | -0.0212 | No |
| 18 | ADK | ADK Entrez, Source | adenosine kinase | 3156 | 0.067 | -0.0178 | No |
| 19 | ARF1 | ARF1 Entrez, Source | ADP-ribosylation factor 1 | 3732 | 0.051 | -0.0561 | No |
| 20 | CUGBP1 | CUGBP1 Entrez, Source | CUG triplet repeat, RNA binding protein 1 | 4049 | 0.043 | -0.0756 | No |
| 21 | WASL | WASL Entrez, Source | Wiskott-Aldrich syndrome-like | 4230 | 0.039 | -0.0854 | No |
| 22 | SNAP25 | SNAP25 Entrez, Source | synaptosomal-associated protein, 25kDa | 4406 | 0.036 | -0.0950 | No |
| 23 | PIGL | PIGL Entrez, Source | phosphatidylinositol glycan anchor biosynthesis, class L | 4560 | 0.033 | -0.1033 | No |
| 24 | ATP5F1 | ATP5F1 Entrez, Source | ATP synthase, H+ transporting, mitochondrial F0 complex, subunit B1 | 4825 | 0.028 | -0.1205 | No |
| 25 | SLC35A4 | SLC35A4 Entrez, Source | solute carrier family 35, member A4 | 4860 | 0.027 | -0.1204 | No |
| 26 | RAD23A | RAD23A Entrez, Source | RAD23 homolog A (S. cerevisiae) | 4946 | 0.025 | -0.1244 | No |
| 27 | TSC1 | TSC1 Entrez, Source | tuberous sclerosis 1 | 4992 | 0.024 | -0.1254 | No |
| 28 | PPP1CC | PPP1CC Entrez, Source | protein phosphatase 1, catalytic subunit, gamma isoform | 4996 | 0.024 | -0.1232 | No |
| 29 | PSMD8 | PSMD8 Entrez, Source | proteasome (prosome, macropain) 26S subunit, non-ATPase, 8 | 5360 | 0.018 | -0.1489 | No |
| 30 | CLK1 | CLK1 Entrez, Source | CDC-like kinase 1 | 5504 | 0.015 | -0.1582 | No |
| 31 | USP48 | USP48 Entrez, Source | ubiquitin specific peptidase 48 | 5600 | 0.013 | -0.1641 | No |
| 32 | ZFP91 | ZFP91 Entrez, Source | zinc finger protein 91 homolog (mouse) | 5690 | 0.011 | -0.1697 | No |
| 33 | ARCN1 | ARCN1 Entrez, Source | archain 1 | 5911 | 0.007 | -0.1856 | No |
| 34 | VAMP2 | VAMP2 Entrez, Source | vesicle-associated membrane protein 2 (synaptobrevin 2) | 6096 | 0.003 | -0.1992 | No |
| 35 | ATF4 | ATF4 Entrez, Source | activating transcription factor 4 (tax-responsive enhancer element B67) | 6295 | -0.000 | -0.2141 | No |
| 36 | TCF4 | TCF4 Entrez, Source | transcription factor 4 | 6338 | -0.001 | -0.2172 | No |
| 37 | USF1 | USF1 Entrez, Source | upstream transcription factor 1 | 6510 | -0.004 | -0.2298 | No |
| 38 | ATP6V0C | ATP6V0C Entrez, Source | ATPase, H+ transporting, lysosomal 16kDa, V0 subunit c | 6588 | -0.005 | -0.2350 | No |
| 39 | NFYC | NFYC Entrez, Source | nuclear transcription factor Y, gamma | 6617 | -0.006 | -0.2366 | No |
| 40 | RAB1A | RAB1A Entrez, Source | RAB1A, member RAS oncogene family | 6729 | -0.008 | -0.2442 | No |
| 41 | ATP5H | ATP5H Entrez, Source | ATP synthase, H+ transporting, mitochondrial F0 complex, subunit d | 7126 | -0.014 | -0.2727 | No |
| 42 | MAEA | MAEA Entrez, Source | macrophage erythroblast attacher | 7146 | -0.015 | -0.2726 | No |
| 43 | NFYA | NFYA Entrez, Source | nuclear transcription factor Y, alpha | 7202 | -0.016 | -0.2752 | No |
| 44 | DAP3 | DAP3 Entrez, Source | death associated protein 3 | 7418 | -0.020 | -0.2894 | No |
| 45 | EIF3S10 | EIF3S10 Entrez, Source | eukaryotic translation initiation factor 3, subunit 10 theta, 150/170kDa | 7771 | -0.027 | -0.3134 | No |
| 46 | ENSA | ENSA Entrez, Source | endosulfine alpha | 8038 | -0.032 | -0.3303 | No |
| 47 | AP3D1 | AP3D1 Entrez, Source | adaptor-related protein complex 3, delta 1 subunit | 8055 | -0.032 | -0.3283 | No |
| 48 | EEF1A1 | EEF1A1 Entrez, Source | eukaryotic translation elongation factor 1 alpha 1 | 8406 | -0.039 | -0.3508 | No |
| 49 | VDAC2 | VDAC2 Entrez, Source | voltage-dependent anion channel 2 | 8408 | -0.040 | -0.3469 | No |
| 50 | PICALM | PICALM Entrez, Source | phosphatidylinositol binding clathrin assembly protein | 8462 | -0.041 | -0.3469 | No |
| 51 | FBXO11 | FBXO11 Entrez, Source | F-box protein 11 | 8731 | -0.046 | -0.3626 | No |
| 52 | RPAP1 | RPAP1 Entrez, Source | RNA polymerase II associated protein 1 | 8923 | -0.050 | -0.3720 | No |
| 53 | NEUROD2 | NEUROD2 Entrez, Source | neurogenic differentiation 2 | 9017 | -0.052 | -0.3738 | No |
| 54 | RAC1 | RAC1 Entrez, Source | ras-related C3 botulinum toxin substrate 1 (rho family, small GTP binding protein Rac1) | 9101 | -0.054 | -0.3748 | No |
| 55 | PIAS3 | PIAS3 Entrez, Source | protein inhibitor of activated STAT, 3 | 9188 | -0.056 | -0.3757 | No |
| 56 | PRPSAP1 | PRPSAP1 Entrez, Source | phosphoribosyl pyrophosphate synthetase-associated protein 1 | 9536 | -0.064 | -0.3956 | No |
| 57 | POLK | POLK Entrez, Source | polymerase (DNA directed) kappa | 9575 | -0.065 | -0.3920 | No |
| 58 | NCOR1 | NCOR1 Entrez, Source | nuclear receptor co-repressor 1 | 9668 | -0.067 | -0.3923 | No |
| 59 | DNAJC7 | DNAJC7 Entrez, Source | DnaJ (Hsp40) homolog, subfamily C, member 7 | 9769 | -0.070 | -0.3930 | No |
| 60 | UBE2D3 | UBE2D3 Entrez, Source | ubiquitin-conjugating enzyme E2D 3 (UBC4/5 homolog, yeast) | 9797 | -0.071 | -0.3880 | No |
| 61 | RFX3 | RFX3 Entrez, Source | regulatory factor X, 3 (influences HLA class II expression) | 10140 | -0.080 | -0.4059 | Yes |
| 62 | RBM5 | RBM5 Entrez, Source | RNA binding motif protein 5 | 10207 | -0.082 | -0.4028 | Yes |
| 63 | MID1 | MID1 Entrez, Source | midline 1 (Opitz/BBB syndrome) | 10356 | -0.086 | -0.4054 | Yes |
| 64 | ATF1 | ATF1 Entrez, Source | activating transcription factor 1 | 10381 | -0.087 | -0.3986 | Yes |
| 65 | PSMB5 | PSMB5 Entrez, Source | proteasome (prosome, macropain) subunit, beta type, 5 | 10521 | -0.091 | -0.4001 | Yes |
| 66 | REST | REST Entrez, Source | RE1-silencing transcription factor | 10648 | -0.095 | -0.4002 | Yes |
| 67 | RPL30 | RPL30 Entrez, Source | ribosomal protein L30 | 10673 | -0.096 | -0.3925 | Yes |
| 68 | POU2F1 | POU2F1 Entrez, Source | POU domain, class 2, transcription factor 1 | 10684 | -0.097 | -0.3837 | Yes |
| 69 | EP300 | EP300 Entrez, Source | E1A binding protein p300 | 10720 | -0.098 | -0.3766 | Yes |
| 70 | EIF4G2 | EIF4G2 Entrez, Source | eukaryotic translation initiation factor 4 gamma, 2 | 10777 | -0.100 | -0.3709 | Yes |
| 71 | APBB3 | APBB3 Entrez, Source | amyloid beta (A4) precursor protein-binding, family B, member 3 | 11051 | -0.110 | -0.3806 | Yes |
| 72 | YY1 | YY1 Entrez, Source | YY1 transcription factor | 11101 | -0.112 | -0.3732 | Yes |
| 73 | RPS27 | RPS27 Entrez, Source | ribosomal protein S27 (metallopanstimulin 1) | 11176 | -0.115 | -0.3674 | Yes |
| 74 | CDH23 | CDH23 Entrez, Source | cadherin-like 23 | 11301 | -0.120 | -0.3648 | Yes |
| 75 | RPL35A | RPL35A Entrez, Source | ribosomal protein L35a | 11499 | -0.129 | -0.3669 | Yes |
| 76 | EIF5 | EIF5 Entrez, Source | eukaryotic translation initiation factor 5 | 11521 | -0.130 | -0.3556 | Yes |
| 77 | GNPAT | GNPAT Entrez, Source | glyceronephosphate O-acyltransferase | 11617 | -0.134 | -0.3494 | Yes |
| 78 | RPA2 | RPA2 Entrez, Source | replication protein A2, 32kDa | 12006 | -0.158 | -0.3631 | Yes |
| 79 | ASB2 | ASB2 Entrez, Source | ankyrin repeat and SOCS box-containing 2 | 12011 | -0.158 | -0.3477 | Yes |
| 80 | PABPN1 | PABPN1 Entrez, Source | poly(A) binding protein, nuclear 1 | 12068 | -0.162 | -0.3359 | Yes |
| 81 | MRPL38 | MRPL38 Entrez, Source | mitochondrial ribosomal protein L38 | 12107 | -0.165 | -0.3223 | Yes |
| 82 | RBM12 | RBM12 Entrez, Source | RNA binding motif protein 12 | 12333 | -0.182 | -0.3213 | Yes |
| 83 | SFRS10 | SFRS10 Entrez, Source | splicing factor, arginine/serine-rich 10 (transformer 2 homolog, Drosophila) | 12334 | -0.182 | -0.3033 | Yes |
| 84 | PRPS1 | PRPS1 Entrez, Source | phosphoribosyl pyrophosphate synthetase 1 | 12339 | -0.183 | -0.2855 | Yes |
| 85 | RALA | RALA Entrez, Source | v-ral simian leukemia viral oncogene homolog A (ras related) | 12488 | -0.197 | -0.2771 | Yes |
| 86 | ATP5G2 | ATP5G2 Entrez, Source | ATP synthase, H+ transporting, mitochondrial F0 complex, subunit C2 (subunit 9) | 12683 | -0.222 | -0.2698 | Yes |
| 87 | PTBP2 | PTBP2 Entrez, Source | polypyrimidine tract binding protein 2 | 12789 | -0.239 | -0.2540 | Yes |
| 88 | RBM3 | RBM3 Entrez, Source | RNA binding motif (RNP1, RRM) protein 3 | 12895 | -0.266 | -0.2356 | Yes |
| 89 | SFPQ | SFPQ Entrez, Source | splicing factor proline/glutamine-rich (polypyrimidine tract binding protein associated) | 12901 | -0.267 | -0.2095 | Yes |
| 90 | NASP | NASP Entrez, Source | nuclear autoantigenic sperm protein (histone-binding) | 12994 | -0.291 | -0.1876 | Yes |
| 91 | CSAD | CSAD Entrez, Source | cysteine sulfinic acid decarboxylase | 13026 | -0.301 | -0.1601 | Yes |
| 92 | HNRPA1 | HNRPA1 Entrez, Source | heterogeneous nuclear ribonucleoprotein A1 | 13080 | -0.318 | -0.1326 | Yes |
| 93 | ODF2 | ODF2 Entrez, Source | outer dense fiber of sperm tails 2 | 13146 | -0.356 | -0.1023 | Yes |
| 94 | ARNT | ARNT Entrez, Source | aryl hydrocarbon receptor nuclear translocator | 13157 | -0.367 | -0.0666 | Yes |
| 95 | H1F0 | H1F0 Entrez, Source | H1 histone family, member 0 | 13165 | -0.372 | -0.0303 | Yes |
| 96 | HNRPD | HNRPD Entrez, Source | heterogeneous nuclear ribonucleoprotein D (AU-rich element RNA binding protein 1, 37kDa) | 13227 | -0.440 | 0.0087 | Yes |