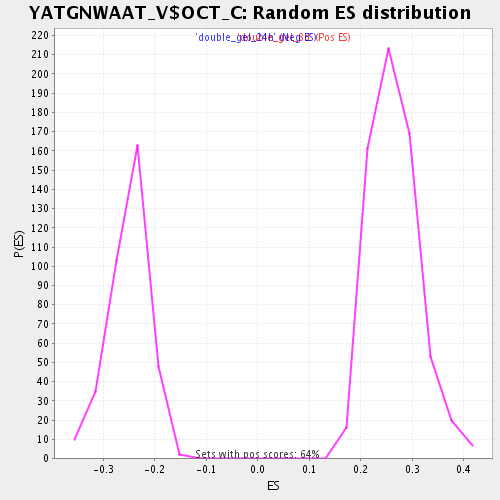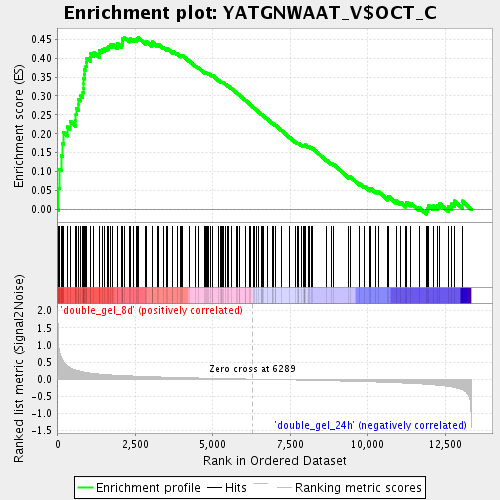
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | double_gel_and_single_gel_rma_expression_values_collapsed_to_symbols.class.cls #double_gel_8d_versus_double_gel_24h.class.cls #double_gel_8d_versus_double_gel_24h_repos |
| Phenotype | class.cls#double_gel_8d_versus_double_gel_24h_repos |
| Upregulated in class | double_gel_8d |
| GeneSet | YATGNWAAT_V$OCT_C |
| Enrichment Score (ES) | 0.455344 |
| Normalized Enrichment Score (NES) | 1.712001 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.051638614 |
| FWER p-Value | 1.0 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | MAF | MAF Entrez, Source | v-maf musculoaponeurotic fibrosarcoma oncogene homolog (avian) | 31 | 0.935 | 0.0560 | Yes |
| 2 | PFKFB1 | PFKFB1 Entrez, Source | 6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 1 | 47 | 0.829 | 0.1065 | Yes |
| 3 | LPL | LPL Entrez, Source | lipoprotein lipase | 107 | 0.646 | 0.1424 | Yes |
| 4 | SLC24A3 | SLC24A3 Entrez, Source | solute carrier family 24 (sodium/potassium/calcium exchanger), member 3 | 151 | 0.563 | 0.1743 | Yes |
| 5 | ALDH1A1 | ALDH1A1 Entrez, Source | aldehyde dehydrogenase 1 family, member A1 | 185 | 0.510 | 0.2036 | Yes |
| 6 | EGR2 | EGR2 Entrez, Source | early growth response 2 (Krox-20 homolog, Drosophila) | 312 | 0.390 | 0.2183 | Yes |
| 7 | CCND1 | CCND1 Entrez, Source | cyclin D1 | 391 | 0.341 | 0.2337 | Yes |
| 8 | GPC4 | GPC4 Entrez, Source | glypican 4 | 577 | 0.262 | 0.2360 | Yes |
| 9 | NFIA | NFIA Entrez, Source | nuclear factor I/A | 581 | 0.262 | 0.2521 | Yes |
| 10 | PPP2R3A | PPP2R3A Entrez, Source | protein phosphatase 2 (formerly 2A), regulatory subunit B'', alpha | 603 | 0.257 | 0.2665 | Yes |
| 11 | LPHN1 | LPHN1 Entrez, Source | latrophilin 1 | 667 | 0.242 | 0.2769 | Yes |
| 12 | SLC10A2 | SLC10A2 Entrez, Source | solute carrier family 10 (sodium/bile acid cotransporter family), member 2 | 671 | 0.242 | 0.2917 | Yes |
| 13 | RAB3C | RAB3C Entrez, Source | RAB3C, member RAS oncogene family | 739 | 0.229 | 0.3009 | Yes |
| 14 | CUTL1 | CUTL1 Entrez, Source | cut-like 1, CCAAT displacement protein (Drosophila) | 807 | 0.212 | 0.3091 | Yes |
| 15 | LMO4 | LMO4 Entrez, Source | LIM domain only 4 | 824 | 0.209 | 0.3209 | Yes |
| 16 | NGFRAP1 | NGFRAP1 Entrez, Source | nerve growth factor receptor (TNFRSF16) associated protein 1 | 830 | 0.208 | 0.3335 | Yes |
| 17 | FOXP1 | FOXP1 Entrez, Source | forkhead box P1 | 833 | 0.207 | 0.3462 | Yes |
| 18 | MAP1B | MAP1B Entrez, Source | microtubule-associated protein 1B | 854 | 0.203 | 0.3574 | Yes |
| 19 | MRAS | MRAS Entrez, Source | muscle RAS oncogene homolog | 863 | 0.201 | 0.3694 | Yes |
| 20 | ADORA1 | ADORA1 Entrez, Source | adenosine A1 receptor | 901 | 0.195 | 0.3787 | Yes |
| 21 | ACTG2 | ACTG2 Entrez, Source | actin, gamma 2, smooth muscle, enteric | 919 | 0.192 | 0.3895 | Yes |
| 22 | IGSF4 | IGSF4 Entrez, Source | immunoglobulin superfamily, member 4 | 932 | 0.190 | 0.4004 | Yes |
| 23 | PITX2 | PITX2 Entrez, Source | paired-like homeodomain transcription factor 2 | 1047 | 0.177 | 0.4028 | Yes |
| 24 | CYR61 | CYR61 Entrez, Source | cysteine-rich, angiogenic inducer, 61 | 1062 | 0.175 | 0.4126 | Yes |
| 25 | DPYSL3 | DPYSL3 Entrez, Source | dihydropyrimidinase-like 3 | 1163 | 0.164 | 0.4153 | Yes |
| 26 | FGF12 | FGF12 Entrez, Source | fibroblast growth factor 12 | 1334 | 0.148 | 0.4117 | Yes |
| 27 | POLM | POLM Entrez, Source | polymerase (DNA directed), mu | 1341 | 0.148 | 0.4204 | Yes |
| 28 | ZFHX1B | ZFHX1B Entrez, Source | zinc finger homeobox 1b | 1427 | 0.140 | 0.4227 | Yes |
| 29 | SCRN1 | SCRN1 Entrez, Source | secernin 1 | 1507 | 0.134 | 0.4251 | Yes |
| 30 | NUDT3 | NUDT3 Entrez, Source | nudix (nucleoside diphosphate linked moiety X)-type motif 3 | 1587 | 0.129 | 0.4272 | Yes |
| 31 | FKRP | FKRP Entrez, Source | fukutin related protein | 1635 | 0.126 | 0.4315 | Yes |
| 32 | RARB | RARB Entrez, Source | retinoic acid receptor, beta | 1690 | 0.123 | 0.4351 | Yes |
| 33 | TRDN | TRDN Entrez, Source | triadin | 1767 | 0.118 | 0.4367 | Yes |
| 34 | NUDT6 | NUDT6 Entrez, Source | nudix (nucleoside diphosphate linked moiety X)-type motif 6 | 1921 | 0.111 | 0.4320 | Yes |
| 35 | CACNG2 | CACNG2 Entrez, Source | calcium channel, voltage-dependent, gamma subunit 2 | 1930 | 0.110 | 0.4383 | Yes |
| 36 | RNASE4 | RNASE4 Entrez, Source | ribonuclease, RNase A family, 4 | 2064 | 0.105 | 0.4347 | Yes |
| 37 | RHOB | RHOB Entrez, Source | ras homolog gene family, member B | 2072 | 0.105 | 0.4408 | Yes |
| 38 | TAS2R7 | TAS2R7 Entrez, Source | taste receptor, type 2, member 7 | 2082 | 0.104 | 0.4466 | Yes |
| 39 | TBXAS1 | TBXAS1 Entrez, Source | thromboxane A synthase 1 (platelet, cytochrome P450, family 5, subfamily A) | 2089 | 0.104 | 0.4526 | Yes |
| 40 | POU1F1 | POU1F1 Entrez, Source | POU domain, class 1, transcription factor 1 (Pit1, growth hormone factor 1) | 2138 | 0.102 | 0.4553 | Yes |
| 41 | CSF3 | CSF3 Entrez, Source | colony stimulating factor 3 (granulocyte) | 2306 | 0.095 | 0.4486 | No |
| 42 | SFRP1 | SFRP1 Entrez, Source | secreted frizzled-related protein 1 | 2327 | 0.094 | 0.4530 | No |
| 43 | EGLN2 | EGLN2 Entrez, Source | egl nine homolog 2 (C. elegans) | 2444 | 0.090 | 0.4498 | No |
| 44 | FZD4 | FZD4 Entrez, Source | frizzled homolog 4 (Drosophila) | 2528 | 0.087 | 0.4489 | No |
| 45 | ITPR3 | ITPR3 Entrez, Source | inositol 1,4,5-triphosphate receptor, type 3 | 2561 | 0.085 | 0.4518 | No |
| 46 | ING1 | ING1 Entrez, Source | inhibitor of growth family, member 1 | 2587 | 0.084 | 0.4551 | No |
| 47 | OTX2 | OTX2 Entrez, Source | orthodenticle homolog 2 (Drosophila) | 2831 | 0.076 | 0.4414 | No |
| 48 | GNB3 | GNB3 Entrez, Source | guanine nucleotide binding protein (G protein), beta polypeptide 3 | 2846 | 0.076 | 0.4451 | No |
| 49 | LMO3 | LMO3 Entrez, Source | LIM domain only 3 (rhombotin-like 2) | 3037 | 0.070 | 0.4351 | No |
| 50 | ERG | ERG Entrez, Source | v-ets erythroblastosis virus E26 oncogene homolog (avian) | 3040 | 0.070 | 0.4393 | No |
| 51 | SYT1 | SYT1 Entrez, Source | synaptotagmin I | 3045 | 0.070 | 0.4434 | No |
| 52 | GRIN3A | GRIN3A Entrez, Source | glutamate receptor, ionotropic, N-methyl-D-aspartate 3A | 3216 | 0.065 | 0.4345 | No |
| 53 | SFRP2 | SFRP2 Entrez, Source | secreted frizzled-related protein 2 | 3247 | 0.064 | 0.4363 | No |
| 54 | ATP1B2 | ATP1B2 Entrez, Source | ATPase, Na+/K+ transporting, beta 2 polypeptide | 3398 | 0.060 | 0.4287 | No |
| 55 | POU2F3 | POU2F3 Entrez, Source | POU domain, class 2, transcription factor 3 | 3508 | 0.057 | 0.4240 | No |
| 56 | FGF20 | FGF20 Entrez, Source | fibroblast growth factor 20 | 3530 | 0.057 | 0.4259 | No |
| 57 | CRISP1 | CRISP1 Entrez, Source | cysteine-rich secretory protein 1 | 3706 | 0.052 | 0.4159 | No |
| 58 | ALDH1A2 | ALDH1A2 Entrez, Source | aldehyde dehydrogenase 1 family, member A2 | 3710 | 0.052 | 0.4189 | No |
| 59 | POU3F4 | POU3F4 Entrez, Source | POU domain, class 3, transcription factor 4 | 3843 | 0.049 | 0.4119 | No |
| 60 | KCNIP4 | KCNIP4 Entrez, Source | Kv channel interacting protein 4 | 3962 | 0.045 | 0.4058 | No |
| 61 | PHC2 | PHC2 Entrez, Source | polyhomeotic homolog 2 (Drosophila) | 3972 | 0.045 | 0.4080 | No |
| 62 | SH3GL3 | SH3GL3 Entrez, Source | SH3-domain GRB2-like 3 | 4015 | 0.044 | 0.4075 | No |
| 63 | HPCAL4 | HPCAL4 Entrez, Source | hippocalcin like 4 | 4243 | 0.039 | 0.3927 | No |
| 64 | GBX2 | GBX2 Entrez, Source | gastrulation brain homeobox 2 | 4453 | 0.035 | 0.3791 | No |
| 65 | FZD2 | FZD2 Entrez, Source | frizzled homolog 2 (Drosophila) | 4530 | 0.033 | 0.3754 | No |
| 66 | SLC6A15 | SLC6A15 Entrez, Source | solute carrier family 6, member 15 | 4739 | 0.029 | 0.3615 | No |
| 67 | NR4A3 | NR4A3 Entrez, Source | nuclear receptor subfamily 4, group A, member 3 | 4753 | 0.029 | 0.3623 | No |
| 68 | ARHGAP4 | ARHGAP4 Entrez, Source | Rho GTPase activating protein 4 | 4793 | 0.028 | 0.3611 | No |
| 69 | RAB26 | RAB26 Entrez, Source | RAB26, member RAS oncogene family | 4823 | 0.028 | 0.3606 | No |
| 70 | PLCB2 | PLCB2 Entrez, Source | phospholipase C, beta 2 | 4867 | 0.027 | 0.3590 | No |
| 71 | TEC | TEC Entrez, Source | tec protein tyrosine kinase | 4933 | 0.025 | 0.3556 | No |
| 72 | THRA | THRA Entrez, Source | thyroid hormone receptor, alpha (erythroblastic leukemia viral (v-erb-a) oncogene homolog, avian) | 4984 | 0.024 | 0.3534 | No |
| 73 | TSC1 | TSC1 Entrez, Source | tuberous sclerosis 1 | 4992 | 0.024 | 0.3544 | No |
| 74 | ACBD4 | ACBD4 Entrez, Source | acyl-Coenzyme A binding domain containing 4 | 5190 | 0.020 | 0.3407 | No |
| 75 | DLX1 | DLX1 Entrez, Source | distal-less homeobox 1 | 5243 | 0.020 | 0.3380 | No |
| 76 | NOS1 | NOS1 Entrez, Source | nitric oxide synthase 1 (neuronal) | 5282 | 0.019 | 0.3363 | No |
| 77 | JUND | JUND Entrez, Source | jun D proto-oncogene | 5289 | 0.019 | 0.3370 | No |
| 78 | DGKG | DGKG Entrez, Source | diacylglycerol kinase, gamma 90kDa | 5313 | 0.019 | 0.3364 | No |
| 79 | COL23A1 | COL23A1 Entrez, Source | collagen, type XXIII, alpha 1 | 5329 | 0.018 | 0.3364 | No |
| 80 | GPR4 | GPR4 Entrez, Source | G protein-coupled receptor 4 | 5412 | 0.016 | 0.3312 | No |
| 81 | NEUROG1 | NEUROG1 Entrez, Source | neurogenin 1 | 5488 | 0.015 | 0.3265 | No |
| 82 | ISL1 | ISL1 Entrez, Source | ISL1 transcription factor, LIM/homeodomain, (islet-1) | 5512 | 0.014 | 0.3256 | No |
| 83 | TMEM24 | TMEM24 Entrez, Source | transmembrane protein 24 | 5591 | 0.013 | 0.3205 | No |
| 84 | NRXN3 | NRXN3 Entrez, Source | neurexin 3 | 5770 | 0.010 | 0.3076 | No |
| 85 | CNOT2 | CNOT2 Entrez, Source | CCR4-NOT transcription complex, subunit 2 | 5786 | 0.009 | 0.3071 | No |
| 86 | CELSR2 | CELSR2 Entrez, Source | cadherin, EGF LAG seven-pass G-type receptor 2 (flamingo homolog, Drosophila) | 5849 | 0.008 | 0.3029 | No |
| 87 | CD79B | CD79B Entrez, Source | CD79b molecule, immunoglobulin-associated beta | 5871 | 0.008 | 0.3018 | No |
| 88 | YWHAB | YWHAB Entrez, Source | tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, beta polypeptide | 6058 | 0.004 | 0.2879 | No |
| 89 | LRRN1 | LRRN1 Entrez, Source | leucine rich repeat neuronal 1 | 6069 | 0.004 | 0.2874 | No |
| 90 | KCNN3 | KCNN3 Entrez, Source | potassium intermediate/small conductance calcium-activated channel, subfamily N, member 3 | 6198 | 0.001 | 0.2778 | No |
| 91 | PCYT1B | PCYT1B Entrez, Source | phosphate cytidylyltransferase 1, choline, beta | 6229 | 0.001 | 0.2756 | No |
| 92 | SLC17A7 | SLC17A7 Entrez, Source | solute carrier family 17 (sodium-dependent inorganic phosphate cotransporter), member 7 | 6323 | -0.000 | 0.2685 | No |
| 93 | TCF4 | TCF4 Entrez, Source | transcription factor 4 | 6338 | -0.001 | 0.2675 | No |
| 94 | BACE2 | BACE2 Entrez, Source | beta-site APP-cleaving enzyme 2 | 6423 | -0.002 | 0.2613 | No |
| 95 | TNNI1 | TNNI1 Entrez, Source | troponin I type 1 (skeletal, slow) | 6476 | -0.003 | 0.2575 | No |
| 96 | IL17F | IL17F Entrez, Source | interleukin 17F | 6554 | -0.005 | 0.2520 | No |
| 97 | ANK3 | ANK3 Entrez, Source | ankyrin 3, node of Ranvier (ankyrin G) | 6582 | -0.005 | 0.2503 | No |
| 98 | ATP6V0C | ATP6V0C Entrez, Source | ATPase, H+ transporting, lysosomal 16kDa, V0 subunit c | 6588 | -0.005 | 0.2502 | No |
| 99 | NRXN1 | NRXN1 Entrez, Source | neurexin 1 | 6602 | -0.006 | 0.2496 | No |
| 100 | PRRX1 | PRRX1 Entrez, Source | paired related homeobox 1 | 6636 | -0.006 | 0.2475 | No |
| 101 | PANK4 | PANK4 Entrez, Source | pantothenate kinase 4 | 6748 | -0.008 | 0.2396 | No |
| 102 | NRAS | NRAS Entrez, Source | neuroblastoma RAS viral (v-ras) oncogene homolog | 6918 | -0.011 | 0.2274 | No |
| 103 | SFRS15 | SFRS15 Entrez, Source | splicing factor, arginine/serine-rich 15 | 6931 | -0.011 | 0.2272 | No |
| 104 | SEMA6C | SEMA6C Entrez, Source | sema domain, transmembrane domain (TM), and cytoplasmic domain, (semaphorin) 6C | 6955 | -0.011 | 0.2262 | No |
| 105 | DLL1 | DLL1 Entrez, Source | delta-like 1 (Drosophila) | 7008 | -0.012 | 0.2230 | No |
| 106 | TOP1 | TOP1 Entrez, Source | topoisomerase (DNA) I | 7017 | -0.013 | 0.2232 | No |
| 107 | DCN | DCN Entrez, Source | decorin | 7217 | -0.016 | 0.2091 | No |
| 108 | GRID2 | GRID2 Entrez, Source | glutamate receptor, ionotropic, delta 2 | 7487 | -0.021 | 0.1901 | No |
| 109 | GDNF | GDNF Entrez, Source | glial cell derived neurotrophic factor | 7663 | -0.025 | 0.1784 | No |
| 110 | PAX6 | PAX6 Entrez, Source | paired box gene 6 (aniridia, keratitis) | 7732 | -0.026 | 0.1748 | No |
| 111 | GCM1 | GCM1 Entrez, Source | glial cells missing homolog 1 (Drosophila) | 7750 | -0.026 | 0.1752 | No |
| 112 | DLGAP4 | DLGAP4 Entrez, Source | discs, large (Drosophila) homolog-associated protein 4 | 7775 | -0.027 | 0.1750 | No |
| 113 | ALK | ALK Entrez, Source | anaplastic lymphoma kinase (Ki-1) | 7870 | -0.029 | 0.1697 | No |
| 114 | DLL3 | DLL3 Entrez, Source | delta-like 3 (Drosophila) | 7929 | -0.030 | 0.1671 | No |
| 115 | TCF2 | TCF2 Entrez, Source | transcription factor 2, hepatic; LF-B3; variant hepatic nuclear factor | 7953 | -0.030 | 0.1673 | No |
| 116 | XYLT2 | XYLT2 Entrez, Source | xylosyltransferase II | 7962 | -0.030 | 0.1686 | No |
| 117 | DSCR1 | DSCR1 Entrez, Source | Down syndrome critical region gene 1 | 7972 | -0.031 | 0.1698 | No |
| 118 | SLC18A3 | SLC18A3 Entrez, Source | solute carrier family 18 (vesicular acetylcholine), member 3 | 7984 | -0.031 | 0.1709 | No |
| 119 | ESRRA | ESRRA Entrez, Source | estrogen-related receptor alpha | 8092 | -0.033 | 0.1649 | No |
| 120 | GPR85 | GPR85 Entrez, Source | G protein-coupled receptor 85 | 8109 | -0.034 | 0.1658 | No |
| 121 | CDK2 | CDK2 Entrez, Source | cyclin-dependent kinase 2 | 8169 | -0.035 | 0.1635 | No |
| 122 | RAB5A | RAB5A Entrez, Source | RAB5A, member RAS oncogene family | 8227 | -0.036 | 0.1614 | No |
| 123 | AMBN | AMBN Entrez, Source | ameloblastin (enamel matrix protein) | 8671 | -0.045 | 0.1306 | No |
| 124 | NXPH1 | NXPH1 Entrez, Source | neurexophilin 1 | 8833 | -0.049 | 0.1214 | No |
| 125 | ADORA2A | ADORA2A Entrez, Source | adenosine A2a receptor | 8894 | -0.050 | 0.1200 | No |
| 126 | H3F3B | H3F3B Entrez, Source | H3 histone, family 3B (H3.3B) | 9390 | -0.060 | 0.0862 | No |
| 127 | PCDH8 | PCDH8 Entrez, Source | protocadherin 8 | 9427 | -0.061 | 0.0873 | No |
| 128 | SYNPR | SYNPR Entrez, Source | synaptoporin | 9740 | -0.069 | 0.0679 | No |
| 129 | AKT2 | AKT2 Entrez, Source | v-akt murine thymoma viral oncogene homolog 2 | 9889 | -0.073 | 0.0613 | No |
| 130 | CRYZL1 | CRYZL1 Entrez, Source | crystallin, zeta (quinone reductase)-like 1 | 10057 | -0.078 | 0.0535 | No |
| 131 | LHX3 | LHX3 Entrez, Source | LIM homeobox 3 | 10087 | -0.079 | 0.0562 | No |
| 132 | LYN | LYN Entrez, Source | v-yes-1 Yamaguchi sarcoma viral related oncogene homolog | 10258 | -0.083 | 0.0485 | No |
| 133 | MID1 | MID1 Entrez, Source | midline 1 (Opitz/BBB syndrome) | 10356 | -0.086 | 0.0466 | No |
| 134 | REST | REST Entrez, Source | RE1-silencing transcription factor | 10648 | -0.095 | 0.0305 | No |
| 135 | MTUS1 | MTUS1 Entrez, Source | mitochondrial tumor suppressor 1 | 10665 | -0.096 | 0.0352 | No |
| 136 | ITSN1 | ITSN1 Entrez, Source | intersectin 1 (SH3 domain protein) | 10914 | -0.105 | 0.0230 | No |
| 137 | ADM | ADM Entrez, Source | adrenomedullin | 11063 | -0.110 | 0.0187 | No |
| 138 | ID2 | ID2 Entrez, Source | inhibitor of DNA binding 2, dominant negative helix-loop-helix protein | 11229 | -0.117 | 0.0135 | No |
| 139 | TSNAX | TSNAX Entrez, Source | translin-associated factor X | 11254 | -0.118 | 0.0190 | No |
| 140 | TAS2R13 | TAS2R13 Entrez, Source | taste receptor, type 2, member 13 | 11390 | -0.124 | 0.0165 | No |
| 141 | DNAH7 | DNAH7 Entrez, Source | dynein, axonemal, heavy chain 7 | 11659 | -0.136 | 0.0047 | No |
| 142 | ABTB2 | ABTB2 Entrez, Source | ankyrin repeat and BTB (POZ) domain containing 2 | 11886 | -0.149 | -0.0031 | No |
| 143 | PTEN | PTEN Entrez, Source | phosphatase and tensin homolog (mutated in multiple advanced cancers 1) | 11925 | -0.152 | 0.0035 | No |
| 144 | HOXA5 | HOXA5 Entrez, Source | homeobox A5 | 11961 | -0.154 | 0.0104 | No |
| 145 | BAT2 | BAT2 Entrez, Source | HLA-B associated transcript 2 | 12112 | -0.166 | 0.0094 | No |
| 146 | STMN1 | STMN1 Entrez, Source | stathmin 1/oncoprotein 18 | 12244 | -0.175 | 0.0104 | No |
| 147 | ADNP | ADNP Entrez, Source | activity-dependent neuroprotector | 12323 | -0.181 | 0.0158 | No |
| 148 | BZW2 | BZW2 Entrez, Source | basic leucine zipper and W2 domains 2 | 12597 | -0.210 | 0.0082 | No |
| 149 | ARHGEF12 | ARHGEF12 Entrez, Source | Rho guanine nucleotide exchange factor (GEF) 12 | 12699 | -0.225 | 0.0145 | No |
| 150 | ITGB3BP | ITGB3BP Entrez, Source | integrin beta 3 binding protein (beta3-endonexin) | 12793 | -0.240 | 0.0224 | No |
| 151 | BST2 | BST2 Entrez, Source | bone marrow stromal cell antigen 2 | 13047 | -0.306 | 0.0224 | No |