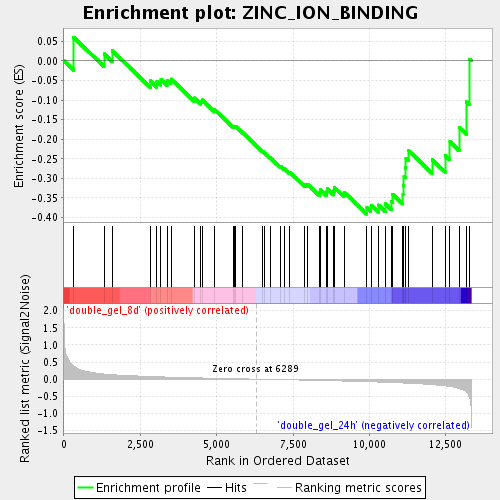
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | double_gel_and_single_gel_rma_expression_values_collapsed_to_symbols.class.cls #double_gel_8d_versus_double_gel_24h.class.cls #double_gel_8d_versus_double_gel_24h_repos |
| Phenotype | class.cls#double_gel_8d_versus_double_gel_24h_repos |
| Upregulated in class | double_gel_24h |
| GeneSet | ZINC_ION_BINDING |
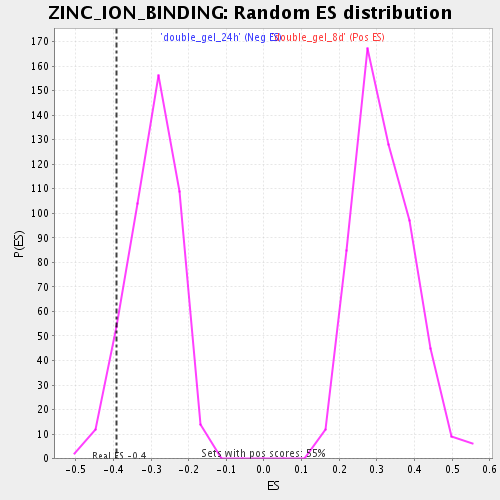
| Enrichment Score (ES) | -0.39125445 |
| Normalized Enrichment Score (NES) | -1.3216354 |
| Nominal p-value | 0.082039915 |
| FDR q-value | 0.38367397 |
| FWER p-Value | 1.0 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | GDA | GDA Entrez, Source | guanine deaminase | 317 | 0.387 | 0.0603 | No |
| 2 | CD4 | CD4 Entrez, Source | CD4 molecule | 1315 | 0.149 | 0.0178 | No |
| 3 | TIMM8B | TIMM8B Entrez, Source | translocase of inner mitochondrial membrane 8 homolog B (yeast) | 1583 | 0.129 | 0.0259 | No |
| 4 | RASSF1 | RASSF1 Entrez, Source | Ras association (RalGDS/AF-6) domain family 1 | 2826 | 0.077 | -0.0509 | No |
| 5 | MEP1B | MEP1B Entrez, Source | meprin A, beta | 3044 | 0.070 | -0.0520 | No |
| 6 | MMP14 | MMP14 Entrez, Source | matrix metallopeptidase 14 (membrane-inserted) | 3176 | 0.066 | -0.0474 | No |
| 7 | MSRB2 | MSRB2 Entrez, Source | methionine sulfoxide reductase B2 | 3376 | 0.061 | -0.0491 | No |
| 8 | LTA4H | LTA4H Entrez, Source | leukotriene A4 hydrolase | 3505 | 0.057 | -0.0462 | No |
| 9 | PEX12 | PEX12 Entrez, Source | peroxisomal biogenesis factor 12 | 4261 | 0.038 | -0.0947 | No |
| 10 | S100B | S100B Entrez, Source | S100 calcium binding protein B | 4471 | 0.035 | -0.1029 | No |
| 11 | CSRP1 | CSRP1 Entrez, Source | cysteine and glycine-rich protein 1 | 4522 | 0.033 | -0.0994 | No |
| 12 | MMP10 | MMP10 Entrez, Source | matrix metallopeptidase 10 (stromelysin 2) | 4938 | 0.025 | -0.1251 | No |
| 13 | MMP2 | MMP2 Entrez, Source | matrix metallopeptidase 2 (gelatinase A, 72kDa gelatinase, 72kDa type IV collagenase) | 5540 | 0.014 | -0.1672 | No |
| 14 | CBLC | CBLC Entrez, Source | Cas-Br-M (murine) ecotropic retroviral transforming sequence c | 5584 | 0.013 | -0.1676 | No |
| 15 | PHEX | PHEX Entrez, Source | phosphate regulating endopeptidase homolog, X-linked (hypophosphatemia, vitamin D resistant rickets) | 5619 | 0.012 | -0.1675 | No |
| 16 | BIRC5 | BIRC5 Entrez, Source | baculoviral IAP repeat-containing 5 (survivin) | 5848 | 0.008 | -0.1829 | No |
| 17 | MMP8 | MMP8 Entrez, Source | matrix metallopeptidase 8 (neutrophil collagenase) | 6500 | -0.004 | -0.2311 | No |
| 18 | SIRT6 | SIRT6 Entrez, Source | sirtuin (silent mating type information regulation 2 homolog) 6 (S. cerevisiae) | 6551 | -0.005 | -0.2338 | No |
| 19 | CBLB | CBLB Entrez, Source | Cas-Br-M (murine) ecotropic retroviral transforming sequence b | 6744 | -0.008 | -0.2465 | No |
| 20 | SOD1 | SOD1 Entrez, Source | superoxide dismutase 1, soluble (amyotrophic lateral sclerosis 1 (adult)) | 7093 | -0.014 | -0.2697 | No |
| 21 | MNAT1 | MNAT1 Entrez, Source | menage a trois homolog 1, cyclin H assembly factor (Xenopus laevis) | 7213 | -0.016 | -0.2751 | No |
| 22 | RPS29 | RPS29 Entrez, Source | ribosomal protein S29 | 7396 | -0.020 | -0.2845 | No |
| 23 | SIRT2 | SIRT2 Entrez, Source | sirtuin (silent mating type information regulation 2 homolog) 2 (S. cerevisiae) | 7882 | -0.029 | -0.3147 | No |
| 24 | TIMM9 | TIMM9 Entrez, Source | translocase of inner mitochondrial membrane 9 homolog (yeast) | 7975 | -0.031 | -0.3149 | No |
| 25 | CTCF | CTCF Entrez, Source | CCCTC-binding factor (zinc finger protein) | 8374 | -0.039 | -0.3364 | No |
| 26 | MMP12 | MMP12 Entrez, Source | matrix metallopeptidase 12 (macrophage elastase) | 8384 | -0.039 | -0.3285 | No |
| 27 | SLU7 | SLU7 Entrez, Source | - | 8593 | -0.043 | -0.3348 | No |
| 28 | MT3 | MT3 Entrez, Source | metallothionein 3 (growth inhibitory factor (neurotrophic)) | 8612 | -0.044 | -0.3266 | No |
| 29 | ECE2 | ECE2 Entrez, Source | endothelin converting enzyme 2 | 8825 | -0.048 | -0.3321 | No |
| 30 | DUSP12 | DUSP12 Entrez, Source | dual specificity phosphatase 12 | 8847 | -0.049 | -0.3230 | No |
| 31 | RNF4 | RNF4 Entrez, Source | ring finger protein 4 | 9168 | -0.055 | -0.3351 | No |
| 32 | LNPEP | LNPEP Entrez, Source | leucyl/cystinyl aminopeptidase | 9916 | -0.074 | -0.3751 | Yes |
| 33 | TIMM10 | TIMM10 Entrez, Source | translocase of inner mitochondrial membrane 10 homolog (yeast) | 10050 | -0.078 | -0.3681 | Yes |
| 34 | MMP9 | MMP9 Entrez, Source | matrix metallopeptidase 9 (gelatinase B, 92kDa gelatinase, 92kDa type IV collagenase) | 10293 | -0.084 | -0.3680 | Yes |
| 35 | PIAS2 | PIAS2 Entrez, Source | protein inhibitor of activated STAT, 2 | 10516 | -0.091 | -0.3650 | Yes |
| 36 | TIMM13 | TIMM13 Entrez, Source | translocase of inner mitochondrial membrane 13 homolog (yeast) | 10719 | -0.098 | -0.3589 | Yes |
| 37 | CA2 | CA2 Entrez, Source | carbonic anhydrase II | 10759 | -0.099 | -0.3402 | Yes |
| 38 | BRCA1 | BRCA1 Entrez, Source | breast cancer 1, early onset | 11096 | -0.112 | -0.3411 | Yes |
| 39 | YY1 | YY1 Entrez, Source | YY1 transcription factor | 11101 | -0.112 | -0.3170 | Yes |
| 40 | MMP16 | MMP16 Entrez, Source | matrix metallopeptidase 16 (membrane-inserted) | 11129 | -0.113 | -0.2944 | Yes |
| 41 | RPS27 | RPS27 Entrez, Source | ribosomal protein S27 (metallopanstimulin 1) | 11176 | -0.115 | -0.2727 | Yes |
| 42 | HDAC10 | HDAC10 Entrez, Source | histone deacetylase 10 | 11193 | -0.116 | -0.2487 | Yes |
| 43 | TP53 | TP53 Entrez, Source | tumor protein p53 (Li-Fraumeni syndrome) | 11266 | -0.119 | -0.2282 | Yes |
| 44 | CHD4 | CHD4 Entrez, Source | chromodomain helicase DNA binding protein 4 | 12048 | -0.161 | -0.2519 | Yes |
| 45 | KPNB1 | KPNB1 Entrez, Source | karyopherin (importin) beta 1 | 12477 | -0.196 | -0.2414 | Yes |
| 46 | APOBEC1 | APOBEC1 Entrez, Source | apolipoprotein B mRNA editing enzyme, catalytic polypeptide 1 | 12633 | -0.215 | -0.2062 | Yes |
| 47 | PRDM4 | PRDM4 Entrez, Source | PR domain containing 4 | 12936 | -0.273 | -0.1695 | Yes |
| 48 | MDM4 | MDM4 Entrez, Source | Mdm4, transformed 3T3 cell double minute 4, p53 binding protein (mouse) | 13174 | -0.380 | -0.1046 | Yes |
| 49 | CRIP2 | CRIP2 Entrez, Source | cysteine-rich protein 2 | 13280 | -0.538 | 0.0047 | Yes |