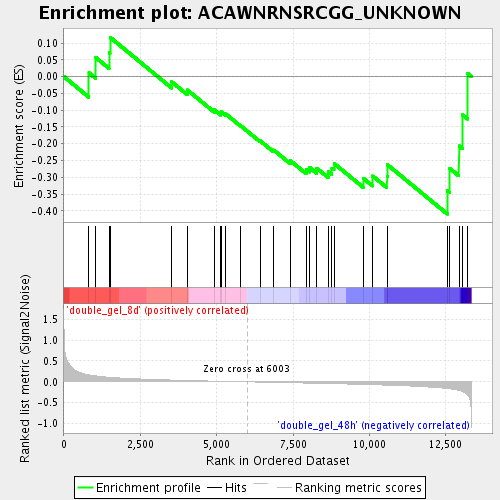
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | double_gel_and_single_gel_rma_expression_values_collapsed_to_symbols.class.cls #double_gel_8d_versus_double_gel_48h.class.cls #double_gel_8d_versus_double_gel_48h_repos |
| Phenotype | class.cls#double_gel_8d_versus_double_gel_48h_repos |
| Upregulated in class | double_gel_48h |
| GeneSet | ACAWNRNSRCGG_UNKNOWN |
| Enrichment Score (ES) | -0.40984857 |
| Normalized Enrichment Score (NES) | -1.2627934 |
| Nominal p-value | 0.15051547 |
| FDR q-value | 0.52626824 |
| FWER p-Value | 1.0 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | CKB | CKB Entrez, Source | creatine kinase, brain | 820 | 0.166 | 0.0114 | No |
| 2 | NR4A1 | NR4A1 Entrez, Source | nuclear receptor subfamily 4, group A, member 1 | 1043 | 0.142 | 0.0570 | No |
| 3 | SMPD3 | SMPD3 Entrez, Source | sphingomyelin phosphodiesterase 3, neutral membrane (neutral sphingomyelinase II) | 1478 | 0.109 | 0.0725 | No |
| 4 | TGFB1I1 | TGFB1I1 Entrez, Source | transforming growth factor beta 1 induced transcript 1 | 1509 | 0.108 | 0.1175 | No |
| 5 | NCOA6 | NCOA6 Entrez, Source | nuclear receptor coactivator 6 | 3527 | 0.042 | -0.0154 | No |
| 6 | ELOVL5 | ELOVL5 Entrez, Source | ELOVL family member 5, elongation of long chain fatty acids (FEN1/Elo2, SUR4/Elo3-like, yeast) | 4037 | 0.032 | -0.0396 | No |
| 7 | NR2C2 | NR2C2 Entrez, Source | nuclear receptor subfamily 2, group C, member 2 | 4923 | 0.016 | -0.0989 | No |
| 8 | SUMO1 | SUMO1 Entrez, Source | SMT3 suppressor of mif two 3 homolog 1 (S. cerevisiae) | 5136 | 0.013 | -0.1092 | No |
| 9 | ARF1 | ARF1 Entrez, Source | ADP-ribosylation factor 1 | 5149 | 0.012 | -0.1047 | No |
| 10 | SLIT2 | SLIT2 Entrez, Source | slit homolog 2 (Drosophila) | 5293 | 0.010 | -0.1109 | No |
| 11 | MTMR3 | MTMR3 Entrez, Source | myotubularin related protein 3 | 5782 | 0.003 | -0.1463 | No |
| 12 | CXXC1 | CXXC1 Entrez, Source | CXXC finger 1 (PHD domain) | 6422 | -0.007 | -0.1914 | No |
| 13 | CTCF | CTCF Entrez, Source | CCCTC-binding factor (zinc finger protein) | 6862 | -0.013 | -0.2187 | No |
| 14 | NFYC | NFYC Entrez, Source | nuclear transcription factor Y, gamma | 7410 | -0.021 | -0.2505 | No |
| 15 | RXRB | RXRB Entrez, Source | retinoid X receptor, beta | 7930 | -0.030 | -0.2765 | No |
| 16 | DDX5 | DDX5 Entrez, Source | DEAD (Asp-Glu-Ala-Asp) box polypeptide 5 | 8037 | -0.031 | -0.2707 | No |
| 17 | DDX17 | DDX17 Entrez, Source | DEAD (Asp-Glu-Ala-Asp) box polypeptide 17 | 8274 | -0.035 | -0.2729 | No |
| 18 | EP300 | EP300 Entrez, Source | E1A binding protein p300 | 8660 | -0.041 | -0.2837 | No |
| 19 | RAGE | RAGE Entrez, Source | renal tumor antigen | 8769 | -0.043 | -0.2728 | No |
| 20 | YWHAE | YWHAE Entrez, Source | tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, epsilon polypeptide | 8841 | -0.044 | -0.2586 | No |
| 21 | EIF4G2 | EIF4G2 Entrez, Source | eukaryotic translation initiation factor 4 gamma, 2 | 9796 | -0.062 | -0.3031 | No |
| 22 | PTBP1 | PTBP1 Entrez, Source | polypyrimidine tract binding protein 1 | 10104 | -0.068 | -0.2961 | No |
| 23 | AKAP8 | AKAP8 Entrez, Source | A kinase (PRKA) anchor protein 8 | 10573 | -0.079 | -0.2965 | No |
| 24 | HES6 | HES6 Entrez, Source | hairy and enhancer of split 6 (Drosophila) | 10589 | -0.080 | -0.2627 | No |
| 25 | SFRS10 | SFRS10 Entrez, Source | splicing factor, arginine/serine-rich 10 (transformer 2 homolog, Drosophila) | 12549 | -0.158 | -0.3404 | Yes |
| 26 | CRKL | CRKL Entrez, Source | v-crk sarcoma virus CT10 oncogene homolog (avian)-like | 12628 | -0.166 | -0.2736 | Yes |
| 27 | RBM3 | RBM3 Entrez, Source | RNA binding motif (RNP1, RRM) protein 3 | 12930 | -0.204 | -0.2066 | Yes |
| 28 | ILF3 | ILF3 Entrez, Source | interleukin enhancer binding factor 3, 90kDa | 13043 | -0.231 | -0.1138 | Yes |
| 29 | SFPQ | SFPQ Entrez, Source | splicing factor proline/glutamine-rich (polypyrimidine tract binding protein associated) | 13213 | -0.310 | 0.0097 | Yes |