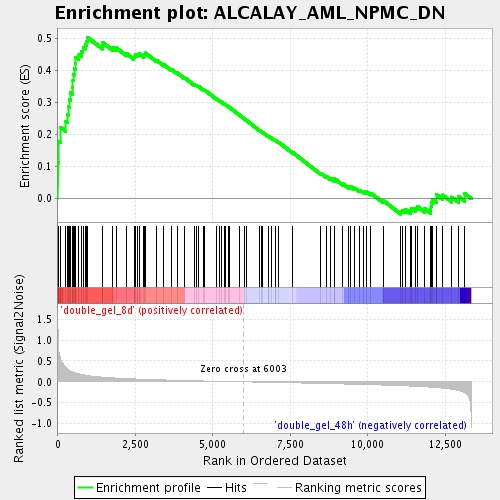
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | double_gel_and_single_gel_rma_expression_values_collapsed_to_symbols.class.cls #double_gel_8d_versus_double_gel_48h.class.cls #double_gel_8d_versus_double_gel_48h_repos |
| Phenotype | class.cls#double_gel_8d_versus_double_gel_48h_repos |
| Upregulated in class | double_gel_8d |
| GeneSet | ALCALAY_AML_NPMC_DN |
| Enrichment Score (ES) | 0.5024176 |
| Normalized Enrichment Score (NES) | 1.7998079 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.025666272 |
| FWER p-Value | 0.971 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | IGFBP7 | IGFBP7 Entrez, Source | insulin-like growth factor binding protein 7 | 2 | 1.254 | 0.1141 | Yes |
| 2 | HLA-DMA | HLA-DMA Entrez, Source | major histocompatibility complex, class II, DM alpha | 32 | 0.725 | 0.1780 | Yes |
| 3 | IL1R2 | IL1R2 Entrez, Source | interleukin 1 receptor, type II | 89 | 0.531 | 0.2221 | Yes |
| 4 | IFITM2 | IFITM2 Entrez, Source | interferon induced transmembrane protein 2 (1-8D) | 254 | 0.339 | 0.2407 | Yes |
| 5 | SPARC | SPARC Entrez, Source | secreted protein, acidic, cysteine-rich (osteonectin) | 317 | 0.293 | 0.2627 | Yes |
| 6 | CD48 | CD48 Entrez, Source | CD48 molecule | 352 | 0.281 | 0.2857 | Yes |
| 7 | RGS10 | RGS10 Entrez, Source | regulator of G-protein signalling 10 | 370 | 0.272 | 0.3093 | Yes |
| 8 | PXMP2 | PXMP2 Entrez, Source | peroxisomal membrane protein 2, 22kDa | 408 | 0.257 | 0.3298 | Yes |
| 9 | ACAA2 | ACAA2 Entrez, Source | acetyl-Coenzyme A acyltransferase 2 (mitochondrial 3-oxoacyl-Coenzyme A thiolase) | 470 | 0.238 | 0.3469 | Yes |
| 10 | GSN | GSN Entrez, Source | gelsolin (amyloidosis, Finnish type) | 483 | 0.233 | 0.3672 | Yes |
| 11 | ITM2C | ITM2C Entrez, Source | integral membrane protein 2C | 491 | 0.231 | 0.3877 | Yes |
| 12 | PROM1 | PROM1 Entrez, Source | prominin 1 | 530 | 0.220 | 0.4049 | Yes |
| 13 | HADH | HADH Entrez, Source | hydroxyacyl-Coenzyme A dehydrogenase | 566 | 0.213 | 0.4216 | Yes |
| 14 | PLAGL1 | PLAGL1 Entrez, Source | pleiomorphic adenoma gene-like 1 | 576 | 0.211 | 0.4401 | Yes |
| 15 | CD96 | CD96 Entrez, Source | CD96 molecule | 677 | 0.189 | 0.4498 | Yes |
| 16 | ITGA9 | ITGA9 Entrez, Source | integrin, alpha 9 | 767 | 0.175 | 0.4590 | Yes |
| 17 | ACAT1 | ACAT1 Entrez, Source | acetyl-Coenzyme A acetyltransferase 1 (acetoacetyl Coenzyme A thiolase) | 809 | 0.168 | 0.4712 | Yes |
| 18 | TRAK2 | TRAK2 Entrez, Source | trafficking protein, kinesin binding 2 | 882 | 0.158 | 0.4802 | Yes |
| 19 | KYNU | KYNU Entrez, Source | kynureninase (L-kynurenine hydrolase) | 934 | 0.153 | 0.4903 | Yes |
| 20 | PBX2 | PBX2 Entrez, Source | pre-B-cell leukemia transcription factor 2 | 956 | 0.151 | 0.5024 | Yes |
| 21 | MLLT3 | MLLT3 Entrez, Source | myeloid/lymphoid or mixed-lineage leukemia (trithorax homolog, Drosophila); translocated to, 3 | 1443 | 0.111 | 0.4759 | No |
| 22 | B4GALT6 | B4GALT6 Entrez, Source | UDP-Gal:betaGlcNAc beta 1,4- galactosyltransferase, polypeptide 6 | 1451 | 0.111 | 0.4854 | No |
| 23 | KIF2C | KIF2C Entrez, Source | kinesin family member 2C | 1773 | 0.094 | 0.4698 | No |
| 24 | GNB5 | GNB5 Entrez, Source | guanine nucleotide binding protein (G protein), beta 5 | 1878 | 0.089 | 0.4701 | No |
| 25 | HPCAL1 | HPCAL1 Entrez, Source | hippocalcin-like 1 | 2224 | 0.076 | 0.4510 | No |
| 26 | INPP4B | INPP4B Entrez, Source | inositol polyphosphate-4-phosphatase, type II, 105kDa | 2455 | 0.069 | 0.4399 | No |
| 27 | FAAH | FAAH Entrez, Source | fatty acid amide hydrolase | 2484 | 0.069 | 0.4440 | No |
| 28 | SEPT9 | SEPT9 Entrez, Source | septin 9 | 2498 | 0.068 | 0.4493 | No |
| 29 | IL2RG | IL2RG Entrez, Source | interleukin 2 receptor, gamma (severe combined immunodeficiency) | 2582 | 0.065 | 0.4489 | No |
| 30 | SERPINI1 | SERPINI1 Entrez, Source | serpin peptidase inhibitor, clade I (neuroserpin), member 1 | 2621 | 0.064 | 0.4519 | No |
| 31 | FKBP1B | FKBP1B Entrez, Source | FK506 binding protein 1B, 12.6 kDa | 2776 | 0.061 | 0.4458 | No |
| 32 | PTTG1 | PTTG1 Entrez, Source | pituitary tumor-transforming 1 | 2806 | 0.060 | 0.4491 | No |
| 33 | BAALC | BAALC Entrez, Source | brain and acute leukemia, cytoplasmic | 2819 | 0.060 | 0.4536 | No |
| 34 | STOM | STOM Entrez, Source | stomatin | 3184 | 0.050 | 0.4307 | No |
| 35 | PIP5K1B | PIP5K1B Entrez, Source | phosphatidylinositol-4-phosphate 5-kinase, type I, beta | 3423 | 0.045 | 0.4168 | No |
| 36 | ANK1 | ANK1 Entrez, Source | ankyrin 1, erythrocytic | 3656 | 0.040 | 0.4029 | No |
| 37 | HMMR | HMMR Entrez, Source | hyaluronan-mediated motility receptor (RHAMM) | 3845 | 0.036 | 0.3920 | No |
| 38 | CD69 | CD69 Entrez, Source | CD69 molecule | 4095 | 0.031 | 0.3760 | No |
| 39 | TRH | TRH Entrez, Source | thyrotropin-releasing hormone | 4399 | 0.025 | 0.3554 | No |
| 40 | RPS6KA2 | RPS6KA2 Entrez, Source | ribosomal protein S6 kinase, 90kDa, polypeptide 2 | 4460 | 0.024 | 0.3531 | No |
| 41 | WASF1 | WASF1 Entrez, Source | WAS protein family, member 1 | 4524 | 0.023 | 0.3504 | No |
| 42 | SERPINF1 | SERPINF1 Entrez, Source | serpin peptidase inhibitor, clade F (alpha-2 antiplasmin, pigment epithelium derived factor), member 1 | 4697 | 0.020 | 0.3393 | No |
| 43 | INSL3 | INSL3 Entrez, Source | insulin-like 3 (Leydig cell) | 4741 | 0.019 | 0.3378 | No |
| 44 | HMGN3 | HMGN3 Entrez, Source | high mobility group nucleosomal binding domain 3 | 5116 | 0.013 | 0.3108 | No |
| 45 | TMEM106C | TMEM106C Entrez, Source | transmembrane protein 106C | 5202 | 0.012 | 0.3054 | No |
| 46 | TNFRSF14 | TNFRSF14 Entrez, Source | tumor necrosis factor receptor superfamily, member 14 (herpesvirus entry mediator) | 5269 | 0.011 | 0.3014 | No |
| 47 | CCNF | CCNF Entrez, Source | cyclin F | 5366 | 0.009 | 0.2950 | No |
| 48 | FOXM1 | FOXM1 Entrez, Source | forkhead box M1 | 5399 | 0.009 | 0.2934 | No |
| 49 | GNG7 | GNG7 Entrez, Source | guanine nucleotide binding protein (G protein), gamma 7 | 5497 | 0.007 | 0.2867 | No |
| 50 | TUBG1 | TUBG1 Entrez, Source | tubulin, gamma 1 | 5540 | 0.007 | 0.2842 | No |
| 51 | ASNA1 | ASNA1 Entrez, Source | arsA arsenite transporter, ATP-binding, homolog 1 (bacterial) | 5871 | 0.002 | 0.2594 | No |
| 52 | KIF4A | KIF4A Entrez, Source | kinesin family member 4A | 6014 | -0.000 | 0.2487 | No |
| 53 | SLC1A4 | SLC1A4 Entrez, Source | solute carrier family 1 (glutamate/neutral amino acid transporter), member 4 | 6096 | -0.002 | 0.2428 | No |
| 54 | CRHBP | CRHBP Entrez, Source | corticotropin releasing hormone binding protein | 6512 | -0.008 | 0.2122 | No |
| 55 | KCNIP2 | KCNIP2 Entrez, Source | Kv channel interacting protein 2 | 6562 | -0.009 | 0.2092 | No |
| 56 | NINJ2 | NINJ2 Entrez, Source | ninjurin 2 | 6616 | -0.009 | 0.2061 | No |
| 57 | CCNB1 | CCNB1 Entrez, Source | cyclin B1 | 6785 | -0.012 | 0.1945 | No |
| 58 | EVL | EVL Entrez, Source | Enah/Vasp-like | 6806 | -0.012 | 0.1941 | No |
| 59 | CIITA | CIITA Entrez, Source | class II, major histocompatibility complex, transactivator | 6886 | -0.013 | 0.1894 | No |
| 60 | CDKN2C | CDKN2C Entrez, Source | cyclin-dependent kinase inhibitor 2C (p18, inhibits CDK4) | 7011 | -0.015 | 0.1814 | No |
| 61 | BIRC5 | BIRC5 Entrez, Source | baculoviral IAP repeat-containing 5 (survivin) | 7015 | -0.015 | 0.1825 | No |
| 62 | CORO2A | CORO2A Entrez, Source | coronin, actin binding protein, 2A | 7122 | -0.017 | 0.1761 | No |
| 63 | CD200 | CD200 Entrez, Source | CD200 molecule | 7585 | -0.024 | 0.1434 | No |
| 64 | OGG1 | OGG1 Entrez, Source | 8-oxoguanine DNA glycosylase | 8478 | -0.039 | 0.0796 | No |
| 65 | EGFL7 | EGFL7 Entrez, Source | EGF-like-domain, multiple 7 | 8671 | -0.041 | 0.0689 | No |
| 66 | CHEK1 | CHEK1 Entrez, Source | CHK1 checkpoint homolog (S. pombe) | 8812 | -0.044 | 0.0623 | No |
| 67 | S100B | S100B Entrez, Source | S100 calcium binding protein B | 8923 | -0.046 | 0.0582 | No |
| 68 | CCNA2 | CCNA2 Entrez, Source | cyclin A2 | 8927 | -0.046 | 0.0622 | No |
| 69 | MECR | MECR Entrez, Source | mitochondrial trans-2-enoyl-CoA reductase | 9194 | -0.050 | 0.0467 | No |
| 70 | CDKN3 | CDKN3 Entrez, Source | cyclin-dependent kinase inhibitor 3 (CDK2-associated dual specificity phosphatase) | 9384 | -0.054 | 0.0373 | No |
| 71 | SRD5A1 | SRD5A1 Entrez, Source | steroid-5-alpha-reductase, alpha polypeptide 1 (3-oxo-5 alpha-steroid delta 4-dehydrogenase alpha 1) | 9458 | -0.055 | 0.0369 | No |
| 72 | KIFC1 | KIFC1 Entrez, Source | kinesin family member C1 | 9575 | -0.057 | 0.0333 | No |
| 73 | RAD23B | RAD23B Entrez, Source | RAD23 homolog B (S. cerevisiae) | 9740 | -0.061 | 0.0265 | No |
| 74 | HGF | HGF Entrez, Source | hepatocyte growth factor (hepapoietin A; scatter factor) | 9857 | -0.063 | 0.0235 | No |
| 75 | CD38 | CD38 Entrez, Source | CD38 molecule | 9966 | -0.066 | 0.0213 | No |
| 76 | POU4F1 | POU4F1 Entrez, Source | POU domain, class 4, transcription factor 1 | 10099 | -0.068 | 0.0176 | No |
| 77 | GAMT | GAMT Entrez, Source | guanidinoacetate N-methyltransferase | 10494 | -0.078 | -0.0051 | No |
| 78 | POLD1 | POLD1 Entrez, Source | polymerase (DNA directed), delta 1, catalytic subunit 125kDa | 11062 | -0.092 | -0.0395 | No |
| 79 | HAAO | HAAO Entrez, Source | 3-hydroxyanthranilate 3,4-dioxygenase | 11121 | -0.094 | -0.0353 | No |
| 80 | MTHFD1 | MTHFD1 Entrez, Source | methylenetetrahydrofolate dehydrogenase (NADP+ dependent) 1, methenyltetrahydrofolate cyclohydrolase, formyltetrahydrofolate synthetase | 11221 | -0.096 | -0.0340 | No |
| 81 | VEGF | VEGF Entrez, Source | vascular endothelial growth factor | 11386 | -0.102 | -0.0372 | No |
| 82 | SYNGR1 | SYNGR1 Entrez, Source | synaptogyrin 1 | 11408 | -0.103 | -0.0294 | No |
| 83 | DHRS3 | DHRS3 Entrez, Source | dehydrogenase/reductase (SDR family) member 3 | 11541 | -0.107 | -0.0296 | No |
| 84 | HNRPD | HNRPD Entrez, Source | heterogeneous nuclear ribonucleoprotein D (AU-rich element RNA binding protein 1, 37kDa) | 11613 | -0.110 | -0.0250 | No |
| 85 | RHD | RHD Entrez, Source | Rh blood group, D antigen | 11838 | -0.119 | -0.0311 | No |
| 86 | CCNB2 | CCNB2 Entrez, Source | cyclin B2 | 12025 | -0.128 | -0.0335 | No |
| 87 | ARL2 | ARL2 Entrez, Source | ADP-ribosylation factor-like 2 | 12040 | -0.128 | -0.0228 | No |
| 88 | NUP210 | NUP210 Entrez, Source | nucleoporin 210kDa | 12052 | -0.129 | -0.0119 | No |
| 89 | NPR3 | NPR3 Entrez, Source | natriuretic peptide receptor C/guanylate cyclase C (atrionatriuretic peptide receptor C) | 12092 | -0.131 | -0.0029 | No |
| 90 | TYMS | TYMS Entrez, Source | thymidylate synthetase | 12217 | -0.137 | 0.0003 | No |
| 91 | CDC20 | CDC20 Entrez, Source | CDC20 cell division cycle 20 homolog (S. cerevisiae) | 12218 | -0.137 | 0.0128 | No |
| 92 | DUT | DUT Entrez, Source | dUTP pyrophosphatase | 12414 | -0.149 | 0.0116 | No |
| 93 | ABCG2 | ABCG2 Entrez, Source | ATP-binding cassette, sub-family G (WHITE), member 2 | 12709 | -0.174 | 0.0053 | No |
| 94 | RBM3 | RBM3 Entrez, Source | RNA binding motif (RNP1, RRM) protein 3 | 12930 | -0.204 | 0.0073 | No |
| 95 | HOMER2 | HOMER2 Entrez, Source | homer homolog 2 (Drosophila) | 13128 | -0.260 | 0.0162 | No |