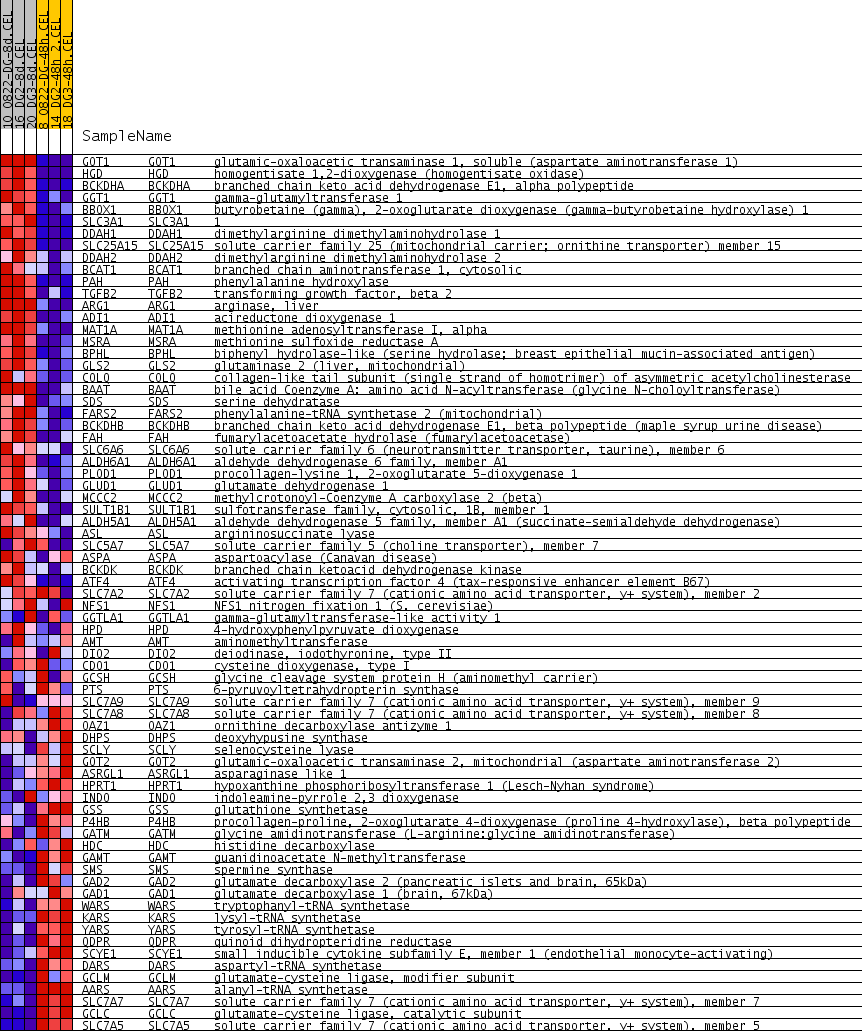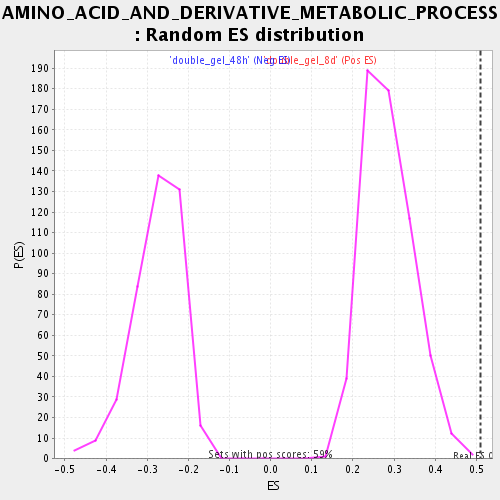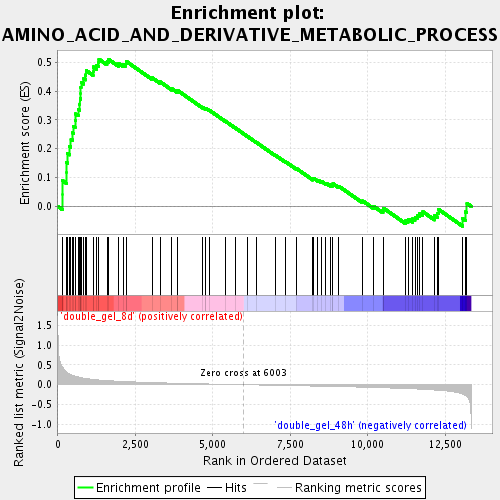
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | double_gel_and_single_gel_rma_expression_values_collapsed_to_symbols.class.cls #double_gel_8d_versus_double_gel_48h.class.cls #double_gel_8d_versus_double_gel_48h_repos |
| Phenotype | class.cls#double_gel_8d_versus_double_gel_48h_repos |
| Upregulated in class | double_gel_8d |
| GeneSet | AMINO_ACID_AND_DERIVATIVE_METABOLIC_PROCESS |
| Enrichment Score (ES) | 0.51051617 |
| Normalized Enrichment Score (NES) | 1.7828372 |
| Nominal p-value | 0.0016977929 |
| FDR q-value | 0.028753242 |
| FWER p-Value | 0.98 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | GOT1 | GOT1 Entrez, Source | glutamic-oxaloacetic transaminase 1, soluble (aspartate aminotransferase 1) | 144 | 0.453 | 0.0396 | Yes |
| 2 | HGD | HGD Entrez, Source | homogentisate 1,2-dioxygenase (homogentisate oxidase) | 146 | 0.449 | 0.0894 | Yes |
| 3 | BCKDHA | BCKDHA Entrez, Source | branched chain keto acid dehydrogenase E1, alpha polypeptide | 272 | 0.326 | 0.1162 | Yes |
| 4 | GGT1 | GGT1 Entrez, Source | gamma-glutamyltransferase 1 | 276 | 0.323 | 0.1520 | Yes |
| 5 | BBOX1 | BBOX1 Entrez, Source | butyrobetaine (gamma), 2-oxoglutarate dioxygenase (gamma-butyrobetaine hydroxylase) 1 | 309 | 0.300 | 0.1829 | Yes |
| 6 | SLC3A1 | SLC3A1 Entrez, Source | 1 | 374 | 0.271 | 0.2082 | Yes |
| 7 | DDAH1 | DDAH1 Entrez, Source | dimethylarginine dimethylaminohydrolase 1 | 421 | 0.254 | 0.2330 | Yes |
| 8 | SLC25A15 | SLC25A15 Entrez, Source | solute carrier family 25 (mitochondrial carrier; ornithine transporter) member 15 | 478 | 0.235 | 0.2550 | Yes |
| 9 | DDAH2 | DDAH2 Entrez, Source | dimethylarginine dimethylaminohydrolase 2 | 501 | 0.227 | 0.2786 | Yes |
| 10 | BCAT1 | BCAT1 Entrez, Source | branched chain aminotransferase 1, cytosolic | 558 | 0.215 | 0.2983 | Yes |
| 11 | PAH | PAH Entrez, Source | phenylalanine hydroxylase | 573 | 0.211 | 0.3208 | Yes |
| 12 | TGFB2 | TGFB2 Entrez, Source | transforming growth factor, beta 2 | 659 | 0.195 | 0.3361 | Yes |
| 13 | ARG1 | ARG1 Entrez, Source | arginase, liver | 704 | 0.185 | 0.3534 | Yes |
| 14 | ADI1 | ADI1 Entrez, Source | acireductone dioxygenase 1 | 712 | 0.183 | 0.3732 | Yes |
| 15 | MAT1A | MAT1A Entrez, Source | methionine adenosyltransferase I, alpha | 734 | 0.180 | 0.3917 | Yes |
| 16 | MSRA | MSRA Entrez, Source | methionine sulfoxide reductase A | 738 | 0.179 | 0.4113 | Yes |
| 17 | BPHL | BPHL Entrez, Source | biphenyl hydrolase-like (serine hydrolase; breast epithelial mucin-associated antigen) | 749 | 0.177 | 0.4302 | Yes |
| 18 | GLS2 | GLS2 Entrez, Source | glutaminase 2 (liver, mitochondrial) | 833 | 0.165 | 0.4423 | Yes |
| 19 | COLQ | COLQ Entrez, Source | collagen-like tail subunit (single strand of homotrimer) of asymmetric acetylcholinesterase | 888 | 0.157 | 0.4558 | Yes |
| 20 | BAAT | BAAT Entrez, Source | bile acid Coenzyme A: amino acid N-acyltransferase (glycine N-choloyltransferase) | 906 | 0.156 | 0.4718 | Yes |
| 21 | SDS | SDS Entrez, Source | serine dehydratase | 1136 | 0.133 | 0.4694 | Yes |
| 22 | FARS2 | FARS2 Entrez, Source | phenylalanine-tRNA synthetase 2 (mitochondrial) | 1142 | 0.133 | 0.4838 | Yes |
| 23 | BCKDHB | BCKDHB Entrez, Source | branched chain keto acid dehydrogenase E1, beta polypeptide (maple syrup urine disease) | 1252 | 0.124 | 0.4893 | Yes |
| 24 | FAH | FAH Entrez, Source | fumarylacetoacetate hydrolase (fumarylacetoacetase) | 1315 | 0.119 | 0.4979 | Yes |
| 25 | SLC6A6 | SLC6A6 Entrez, Source | solute carrier family 6 (neurotransmitter transporter, taurine), member 6 | 1323 | 0.118 | 0.5105 | Yes |
| 26 | ALDH6A1 | ALDH6A1 Entrez, Source | aldehyde dehydrogenase 6 family, member A1 | 1585 | 0.103 | 0.5023 | No |
| 27 | PLOD1 | PLOD1 Entrez, Source | procollagen-lysine 1, 2-oxoglutarate 5-dioxygenase 1 | 1639 | 0.101 | 0.5095 | No |
| 28 | GLUD1 | GLUD1 Entrez, Source | glutamate dehydrogenase 1 | 1962 | 0.087 | 0.4949 | No |
| 29 | MCCC2 | MCCC2 Entrez, Source | methylcrotonoyl-Coenzyme A carboxylase 2 (beta) | 2111 | 0.080 | 0.4927 | No |
| 30 | SULT1B1 | SULT1B1 Entrez, Source | sulfotransferase family, cytosolic, 1B, member 1 | 2197 | 0.077 | 0.4949 | No |
| 31 | ALDH5A1 | ALDH5A1 Entrez, Source | aldehyde dehydrogenase 5 family, member A1 (succinate-semialdehyde dehydrogenase) | 2210 | 0.077 | 0.5026 | No |
| 32 | ASL | ASL Entrez, Source | argininosuccinate lyase | 3041 | 0.053 | 0.4460 | No |
| 33 | SLC5A7 | SLC5A7 Entrez, Source | solute carrier family 5 (choline transporter), member 7 | 3299 | 0.047 | 0.4319 | No |
| 34 | ASPA | ASPA Entrez, Source | aspartoacylase (Canavan disease) | 3681 | 0.039 | 0.4075 | No |
| 35 | BCKDK | BCKDK Entrez, Source | branched chain ketoacid dehydrogenase kinase | 3853 | 0.036 | 0.3986 | No |
| 36 | ATF4 | ATF4 Entrez, Source | activating transcription factor 4 (tax-responsive enhancer element B67) | 3861 | 0.035 | 0.4020 | No |
| 37 | SLC7A2 | SLC7A2 Entrez, Source | solute carrier family 7 (cationic amino acid transporter, y+ system), member 2 | 4675 | 0.020 | 0.3430 | No |
| 38 | NFS1 | NFS1 Entrez, Source | NFS1 nitrogen fixation 1 (S. cerevisiae) | 4759 | 0.019 | 0.3389 | No |
| 39 | GGTLA1 | GGTLA1 Entrez, Source | gamma-glutamyltransferase-like activity 1 | 4761 | 0.019 | 0.3409 | No |
| 40 | HPD | HPD Entrez, Source | 4-hydroxyphenylpyruvate dioxygenase | 4885 | 0.017 | 0.3336 | No |
| 41 | AMT | AMT Entrez, Source | aminomethyltransferase | 5396 | 0.009 | 0.2961 | No |
| 42 | DIO2 | DIO2 Entrez, Source | deiodinase, iodothyronine, type II | 5720 | 0.004 | 0.2722 | No |
| 43 | CDO1 | CDO1 Entrez, Source | cysteine dioxygenase, type I | 6103 | -0.002 | 0.2436 | No |
| 44 | GCSH | GCSH Entrez, Source | glycine cleavage system protein H (aminomethyl carrier) | 6406 | -0.006 | 0.2216 | No |
| 45 | PTS | PTS Entrez, Source | 6-pyruvoyltetrahydropterin synthase | 7010 | -0.015 | 0.1778 | No |
| 46 | SLC7A9 | SLC7A9 Entrez, Source | solute carrier family 7 (cationic amino acid transporter, y+ system), member 9 | 7340 | -0.020 | 0.1553 | No |
| 47 | SLC7A8 | SLC7A8 Entrez, Source | solute carrier family 7 (cationic amino acid transporter, y+ system), member 8 | 7714 | -0.026 | 0.1301 | No |
| 48 | OAZ1 | OAZ1 Entrez, Source | ornithine decarboxylase antizyme 1 | 8231 | -0.034 | 0.0950 | No |
| 49 | DHPS | DHPS Entrez, Source | deoxyhypusine synthase | 8259 | -0.035 | 0.0969 | No |
| 50 | SCLY | SCLY Entrez, Source | selenocysteine lyase | 8389 | -0.037 | 0.0913 | No |
| 51 | GOT2 | GOT2 Entrez, Source | glutamic-oxaloacetic transaminase 2, mitochondrial (aspartate aminotransferase 2) | 8497 | -0.039 | 0.0875 | No |
| 52 | ASRGL1 | ASRGL1 Entrez, Source | asparaginase like 1 | 8649 | -0.041 | 0.0808 | No |
| 53 | HPRT1 | HPRT1 Entrez, Source | hypoxanthine phosphoribosyltransferase 1 (Lesch-Nyhan syndrome) | 8796 | -0.044 | 0.0746 | No |
| 54 | INDO | INDO Entrez, Source | indoleamine-pyrrole 2,3 dioxygenase | 8856 | -0.045 | 0.0751 | No |
| 55 | GSS | GSS Entrez, Source | glutathione synthetase | 8876 | -0.045 | 0.0787 | No |
| 56 | P4HB | P4HB Entrez, Source | procollagen-proline, 2-oxoglutarate 4-dioxygenase (proline 4-hydroxylase), beta polypeptide | 9064 | -0.048 | 0.0700 | No |
| 57 | GATM | GATM Entrez, Source | glycine amidinotransferase (L-arginine:glycine amidinotransferase) | 9836 | -0.063 | 0.0189 | No |
| 58 | HDC | HDC Entrez, Source | histidine decarboxylase | 10195 | -0.070 | -0.0002 | No |
| 59 | GAMT | GAMT Entrez, Source | guanidinoacetate N-methyltransferase | 10494 | -0.078 | -0.0140 | No |
| 60 | SMS | SMS Entrez, Source | spermine synthase | 10501 | -0.078 | -0.0058 | No |
| 61 | GAD2 | GAD2 Entrez, Source | glutamate decarboxylase 2 (pancreatic islets and brain, 65kDa) | 11228 | -0.096 | -0.0498 | No |
| 62 | GAD1 | GAD1 Entrez, Source | glutamate decarboxylase 1 (brain, 67kDa) | 11300 | -0.099 | -0.0442 | No |
| 63 | WARS | WARS Entrez, Source | tryptophanyl-tRNA synthetase | 11445 | -0.104 | -0.0435 | No |
| 64 | KARS | KARS Entrez, Source | lysyl-tRNA synthetase | 11525 | -0.107 | -0.0376 | No |
| 65 | YARS | YARS Entrez, Source | tyrosyl-tRNA synthetase | 11607 | -0.110 | -0.0314 | No |
| 66 | QDPR | QDPR Entrez, Source | quinoid dihydropteridine reductase | 11673 | -0.112 | -0.0239 | No |
| 67 | SCYE1 | SCYE1 Entrez, Source | small inducible cytokine subfamily E, member 1 (endothelial monocyte-activating) | 11779 | -0.116 | -0.0189 | No |
| 68 | DARS | DARS Entrez, Source | aspartyl-tRNA synthetase | 12158 | -0.134 | -0.0324 | No |
| 69 | GCLM | GCLM Entrez, Source | glutamate-cysteine ligase, modifier subunit | 12248 | -0.139 | -0.0236 | No |
| 70 | AARS | AARS Entrez, Source | alanyl-tRNA synthetase | 12282 | -0.141 | -0.0103 | No |
| 71 | SLC7A7 | SLC7A7 Entrez, Source | solute carrier family 7 (cationic amino acid transporter, y+ system), member 7 | 13061 | -0.237 | -0.0425 | No |
| 72 | GCLC | GCLC Entrez, Source | glutamate-cysteine ligase, catalytic subunit | 13151 | -0.271 | -0.0191 | No |
| 73 | SLC7A5 | SLC7A5 Entrez, Source | solute carrier family 7 (cationic amino acid transporter, y+ system), member 5 | 13201 | -0.300 | 0.0106 | No |