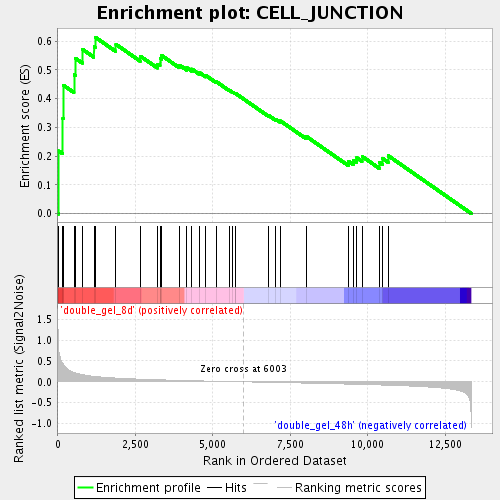
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | double_gel_and_single_gel_rma_expression_values_collapsed_to_symbols.class.cls #double_gel_8d_versus_double_gel_48h.class.cls #double_gel_8d_versus_double_gel_48h_repos |
| Phenotype | class.cls#double_gel_8d_versus_double_gel_48h_repos |
| Upregulated in class | double_gel_8d |
| GeneSet | CELL_JUNCTION |
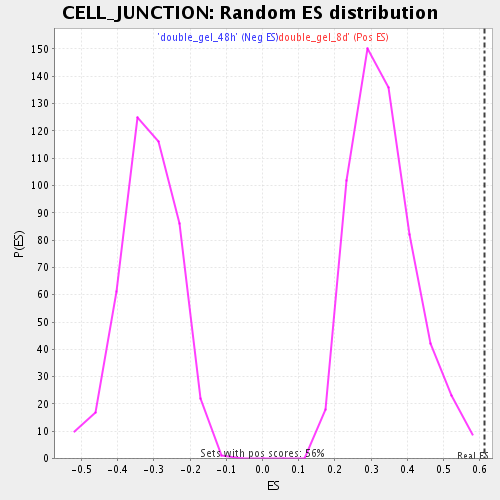
| Enrichment Score (ES) | 0.61390615 |
| Normalized Enrichment Score (NES) | 1.8385293 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.019987125 |
| FWER p-Value | 0.883 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | CLDN7 | CLDN7 Entrez, Source | claudin 7 | 24 | 0.780 | 0.2182 | Yes |
| 2 | CLDN1 | CLDN1 Entrez, Source | claudin 1 | 158 | 0.436 | 0.3313 | Yes |
| 3 | GJA1 | GJA1 Entrez, Source | gap junction protein, alpha 1, 43kDa (connexin 43) | 173 | 0.414 | 0.4472 | Yes |
| 4 | NEXN | NEXN Entrez, Source | nexilin (F actin binding protein) | 520 | 0.223 | 0.4840 | Yes |
| 5 | CLDN4 | CLDN4 Entrez, Source | claudin 4 | 570 | 0.212 | 0.5402 | Yes |
| 6 | CLDN3 | CLDN3 Entrez, Source | claudin 3 | 788 | 0.172 | 0.5723 | Yes |
| 7 | WNK4 | WNK4 Entrez, Source | WNK lysine deficient protein kinase 4 | 1164 | 0.131 | 0.5812 | Yes |
| 8 | PTPRC | PTPRC Entrez, Source | protein tyrosine phosphatase, receptor type, C | 1207 | 0.127 | 0.6139 | Yes |
| 9 | GJB1 | GJB1 Entrez, Source | gap junction protein, beta 1, 32kDa (connexin 32, Charcot-Marie-Tooth neuropathy, X-linked) | 1871 | 0.090 | 0.5894 | No |
| 10 | ZYX | ZYX Entrez, Source | zyxin | 2654 | 0.063 | 0.5486 | No |
| 11 | ACTN1 | ACTN1 Entrez, Source | actinin, alpha 1 | 3213 | 0.049 | 0.5205 | No |
| 12 | CDH1 | CDH1 Entrez, Source | cadherin 1, type 1, E-cadherin (epithelial) | 3296 | 0.047 | 0.5277 | No |
| 13 | ACTN3 | ACTN3 Entrez, Source | actinin, alpha 3 | 3297 | 0.047 | 0.5411 | No |
| 14 | SNIP | SNIP Entrez, Source | - | 3346 | 0.046 | 0.5505 | No |
| 15 | CLDN19 | CLDN19 Entrez, Source | claudin 19 | 3919 | 0.034 | 0.5172 | No |
| 16 | RAB13 | RAB13 Entrez, Source | RAB13, member RAS oncogene family | 4159 | 0.030 | 0.5077 | No |
| 17 | RND1 | RND1 Entrez, Source | Rho family GTPase 1 | 4318 | 0.027 | 0.5034 | No |
| 18 | PARD6A | PARD6A Entrez, Source | par-6 partitioning defective 6 homolog alpha (C.elegans) | 4580 | 0.022 | 0.4900 | No |
| 19 | SORBS3 | SORBS3 Entrez, Source | sorbin and SH3 domain containing 3 | 4774 | 0.019 | 0.4808 | No |
| 20 | BCAR1 | BCAR1 Entrez, Source | breast cancer anti-estrogen resistance 1 | 5112 | 0.013 | 0.4592 | No |
| 21 | CLDN8 | CLDN8 Entrez, Source | claudin 8 | 5544 | 0.007 | 0.4287 | No |
| 22 | GJB2 | GJB2 Entrez, Source | gap junction protein, beta 2, 26kDa (connexin 26) | 5644 | 0.005 | 0.4227 | No |
| 23 | CDC42BPA | CDC42BPA Entrez, Source | CDC42 binding protein kinase alpha (DMPK-like) | 5723 | 0.004 | 0.4179 | No |
| 24 | DLG1 | DLG1 Entrez, Source | discs, large homolog 1 (Drosophila) | 5729 | 0.004 | 0.4186 | No |
| 25 | EVL | EVL Entrez, Source | Enah/Vasp-like | 6806 | -0.012 | 0.3413 | No |
| 26 | CNKSR1 | CNKSR1 Entrez, Source | connector enhancer of kinase suppressor of Ras 1 | 7028 | -0.016 | 0.3290 | No |
| 27 | VAPA | VAPA Entrez, Source | VAMP (vesicle-associated membrane protein)-associated protein A, 33kDa | 7181 | -0.018 | 0.3227 | No |
| 28 | CLDN11 | CLDN11 Entrez, Source | claudin 11 (oligodendrocyte transmembrane protein) | 8011 | -0.031 | 0.2692 | No |
| 29 | PDZD2 | PDZD2 Entrez, Source | PDZ domain containing 2 | 9374 | -0.054 | 0.1820 | No |
| 30 | CLDN14 | CLDN14 Entrez, Source | claudin 14 | 9533 | -0.056 | 0.1860 | No |
| 31 | CLDN5 | CLDN5 Entrez, Source | claudin 5 (transmembrane protein deleted in velocardiofacial syndrome) | 9647 | -0.059 | 0.1941 | No |
| 32 | CDC42BPB | CDC42BPB Entrez, Source | CDC42 binding protein kinase beta (DMPK-like) | 9816 | -0.062 | 0.1991 | No |
| 33 | MLLT4 | MLLT4 Entrez, Source | myeloid/lymphoid or mixed-lineage leukemia (trithorax homolog, Drosophila); translocated to, 4 | 10378 | -0.074 | 0.1779 | No |
| 34 | PARD3 | PARD3 Entrez, Source | par-3 partitioning defective 3 homolog (C. elegans) | 10473 | -0.077 | 0.1925 | No |
| 35 | CLDN16 | CLDN16 Entrez, Source | claudin 16 | 10660 | -0.081 | 0.2015 | No |