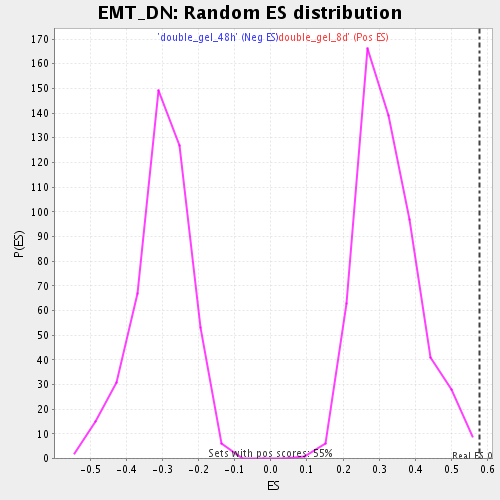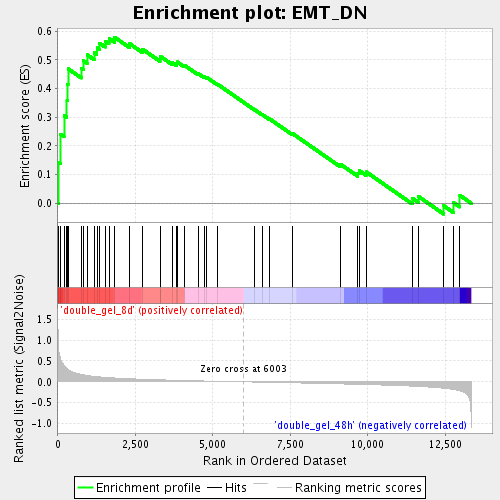
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | double_gel_and_single_gel_rma_expression_values_collapsed_to_symbols.class.cls #double_gel_8d_versus_double_gel_48h.class.cls #double_gel_8d_versus_double_gel_48h_repos |
| Phenotype | class.cls#double_gel_8d_versus_double_gel_48h_repos |
| Upregulated in class | double_gel_8d |
| GeneSet | EMT_DN |
| Enrichment Score (ES) | 0.5790783 |
| Normalized Enrichment Score (NES) | 1.7861471 |
| Nominal p-value | 0.0036363637 |
| FDR q-value | 0.028017437 |
| FWER p-Value | 0.978 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | CTGF | CTGF Entrez, Source | connective tissue growth factor | 27 | 0.757 | 0.1424 | Yes |
| 2 | EGR2 | EGR2 Entrez, Source | early growth response 2 (Krox-20 homolog, Drosophila) | 87 | 0.537 | 0.2405 | Yes |
| 3 | SGK | SGK Entrez, Source | serum/glucocorticoid regulated kinase | 200 | 0.388 | 0.3061 | Yes |
| 4 | PC | PC Entrez, Source | pyruvate carboxylase | 292 | 0.313 | 0.3590 | Yes |
| 5 | ATF3 | ATF3 Entrez, Source | activating transcription factor 3 | 310 | 0.299 | 0.4149 | Yes |
| 6 | TIMP3 | TIMP3 Entrez, Source | TIMP metallopeptidase inhibitor 3 (Sorsby fundus dystrophy, pseudoinflammatory) | 329 | 0.291 | 0.4690 | Yes |
| 7 | EGR1 | EGR1 Entrez, Source | early growth response 1 | 756 | 0.176 | 0.4706 | Yes |
| 8 | ITPR1 | ITPR1 Entrez, Source | inositol 1,4,5-triphosphate receptor, type 1 | 816 | 0.167 | 0.4981 | Yes |
| 9 | F3 | F3 Entrez, Source | coagulation factor III (thromboplastin, tissue factor) | 943 | 0.152 | 0.5176 | Yes |
| 10 | TACSTD1 | TACSTD1 Entrez, Source | tumor-associated calcium signal transducer 1 | 1173 | 0.130 | 0.5253 | Yes |
| 11 | DUSP1 | DUSP1 Entrez, Source | dual specificity phosphatase 1 | 1261 | 0.123 | 0.5422 | Yes |
| 12 | VAMP8 | VAMP8 Entrez, Source | vesicle-associated membrane protein 8 (endobrevin) | 1336 | 0.117 | 0.5590 | Yes |
| 13 | PLK2 | PLK2 Entrez, Source | polo-like kinase 2 (Drosophila) | 1523 | 0.107 | 0.5654 | Yes |
| 14 | TGM2 | TGM2 Entrez, Source | transglutaminase 2 (C polypeptide, protein-glutamine-gamma-glutamyltransferase) | 1650 | 0.100 | 0.5750 | Yes |
| 15 | NUMB | NUMB Entrez, Source | numb homolog (Drosophila) | 1829 | 0.092 | 0.5791 | Yes |
| 16 | SERPINB5 | SERPINB5 Entrez, Source | serpin peptidase inhibitor, clade B (ovalbumin), member 5 | 2312 | 0.074 | 0.5569 | No |
| 17 | CTSH | CTSH Entrez, Source | cathepsin H | 2723 | 0.062 | 0.5379 | No |
| 18 | CDH1 | CDH1 Entrez, Source | cadherin 1, type 1, E-cadherin (epithelial) | 3296 | 0.047 | 0.5040 | No |
| 19 | ID4 | ID4 Entrez, Source | inhibitor of DNA binding 4, dominant negative helix-loop-helix protein | 3304 | 0.047 | 0.5125 | No |
| 20 | TGFB3 | TGFB3 Entrez, Source | transforming growth factor, beta 3 | 3692 | 0.039 | 0.4908 | No |
| 21 | ARHGEF1 | ARHGEF1 Entrez, Source | Rho guanine nucleotide exchange factor (GEF) 1 | 3821 | 0.036 | 0.4881 | No |
| 22 | HMMR | HMMR Entrez, Source | hyaluronan-mediated motility receptor (RHAMM) | 3845 | 0.036 | 0.4932 | No |
| 23 | ATP1A1 | ATP1A1 Entrez, Source | ATPase, Na+/K+ transporting, alpha 1 polypeptide | 4072 | 0.031 | 0.4822 | No |
| 24 | PRKCZ | PRKCZ Entrez, Source | protein kinase C, zeta | 4529 | 0.023 | 0.4522 | No |
| 25 | NRP1 | NRP1 Entrez, Source | neuropilin 1 | 4730 | 0.020 | 0.4409 | No |
| 26 | FBP2 | FBP2 Entrez, Source | fructose-1,6-bisphosphatase 2 | 4805 | 0.018 | 0.4389 | No |
| 27 | CHKA | CHKA Entrez, Source | choline kinase alpha | 5155 | 0.012 | 0.4150 | No |
| 28 | PADI2 | PADI2 Entrez, Source | peptidyl arginine deiminase, type II | 6353 | -0.006 | 0.3261 | No |
| 29 | JUP | JUP Entrez, Source | junction plakoglobin | 6593 | -0.009 | 0.3098 | No |
| 30 | GRB7 | GRB7 Entrez, Source | growth factor receptor-bound protein 7 | 6828 | -0.013 | 0.2946 | No |
| 31 | ID1 | ID1 Entrez, Source | inhibitor of DNA binding 1, dominant negative helix-loop-helix protein | 7557 | -0.023 | 0.2444 | No |
| 32 | TIAM1 | TIAM1 Entrez, Source | T-cell lymphoma invasion and metastasis 1 | 9112 | -0.049 | 0.1369 | No |
| 33 | RARA | RARA Entrez, Source | retinoic acid receptor, alpha | 9679 | -0.059 | 0.1057 | No |
| 34 | STAT5A | STAT5A Entrez, Source | signal transducer and activator of transcription 5A | 9723 | -0.060 | 0.1140 | No |
| 35 | ITGB5 | ITGB5 Entrez, Source | integrin, beta 5 | 9946 | -0.065 | 0.1098 | No |
| 36 | TSC22D1 | TSC22D1 Entrez, Source | TSC22 domain family, member 1 | 11438 | -0.104 | 0.0175 | No |
| 37 | MYH9 | MYH9 Entrez, Source | myosin, heavy chain 9, non-muscle | 11636 | -0.111 | 0.0238 | No |
| 38 | KLF10 | KLF10 Entrez, Source | Kruppel-like factor 10 | 12453 | -0.151 | -0.0086 | No |
| 39 | ID2 | ID2 Entrez, Source | inhibitor of DNA binding 2, dominant negative helix-loop-helix protein | 12777 | -0.181 | 0.0017 | No |
| 40 | CA2 | CA2 Entrez, Source | carbonic anhydrase II | 12972 | -0.213 | 0.0278 | No |