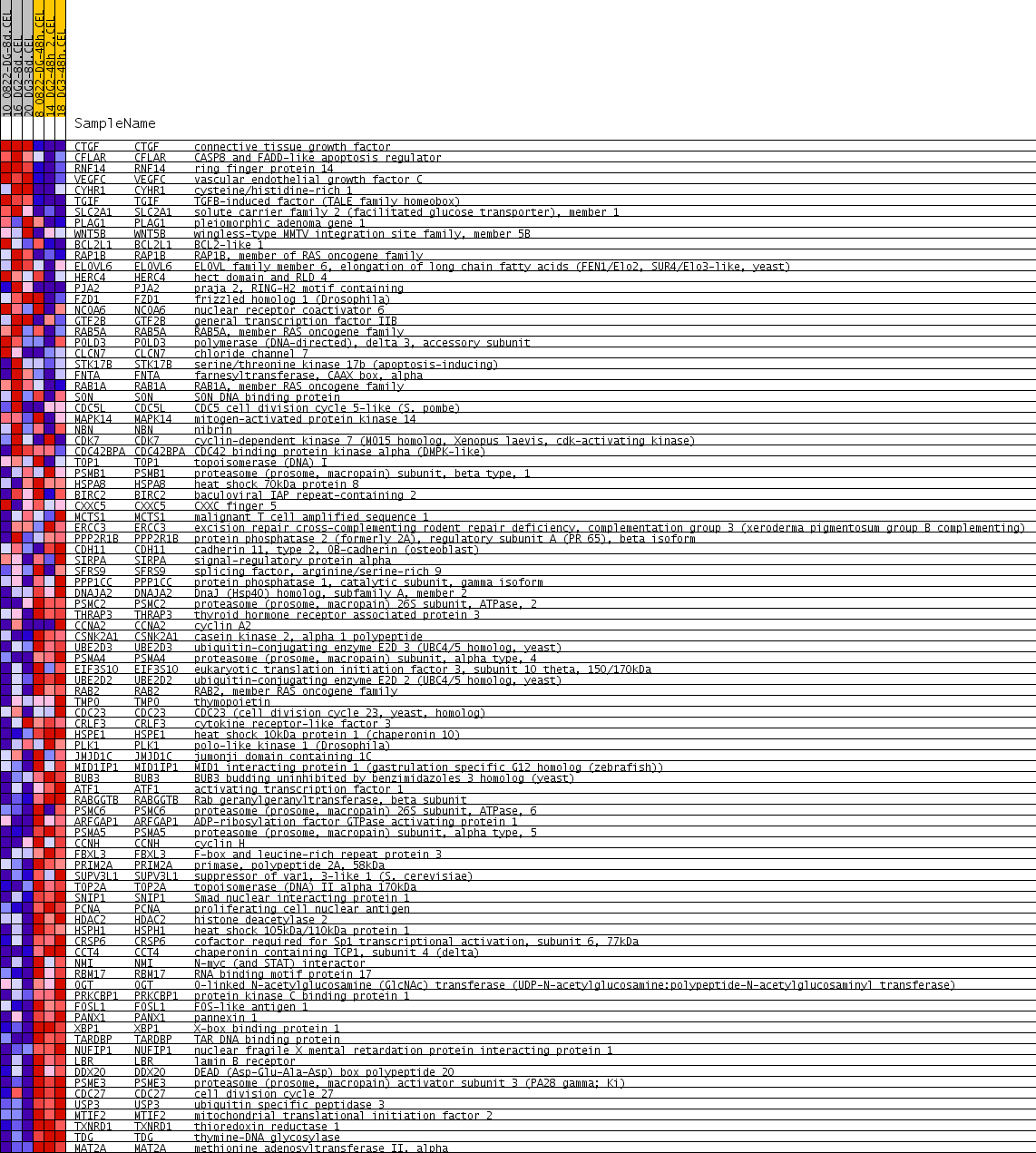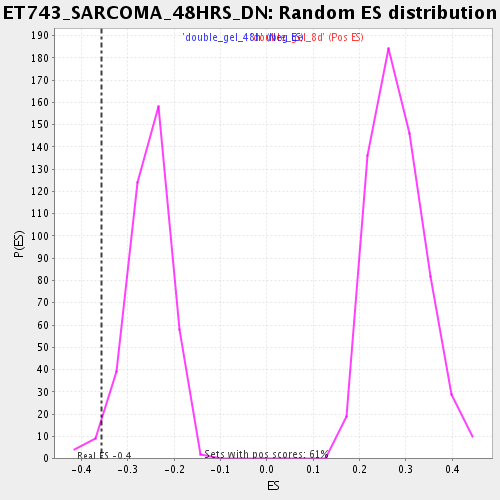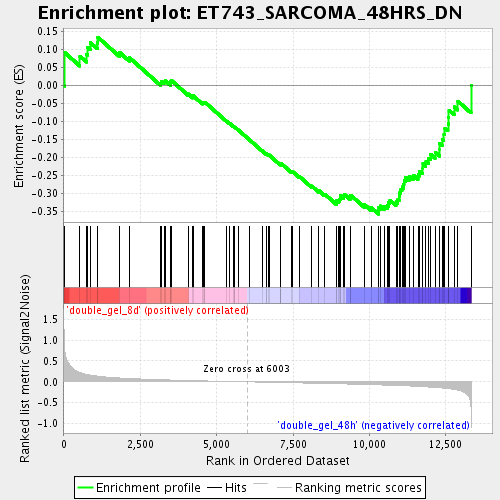
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | double_gel_and_single_gel_rma_expression_values_collapsed_to_symbols.class.cls #double_gel_8d_versus_double_gel_48h.class.cls #double_gel_8d_versus_double_gel_48h_repos |
| Phenotype | class.cls#double_gel_8d_versus_double_gel_48h_repos |
| Upregulated in class | double_gel_48h |
| GeneSet | ET743_SARCOMA_48HRS_DN |
| Enrichment Score (ES) | -0.35711804 |
| Normalized Enrichment Score (NES) | -1.4001167 |
| Nominal p-value | 0.027918782 |
| FDR q-value | 0.34509924 |
| FWER p-Value | 1.0 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | CTGF | CTGF Entrez, Source | connective tissue growth factor | 27 | 0.757 | 0.0912 | No |
| 2 | CFLAR | CFLAR Entrez, Source | CASP8 and FADD-like apoptosis regulator | 523 | 0.222 | 0.0812 | No |
| 3 | RNF14 | RNF14 Entrez, Source | ring finger protein 14 | 740 | 0.178 | 0.0868 | No |
| 4 | VEGFC | VEGFC Entrez, Source | vascular endothelial growth factor C | 778 | 0.173 | 0.1053 | No |
| 5 | CYHR1 | CYHR1 Entrez, Source | cysteine/histidine-rich 1 | 862 | 0.161 | 0.1189 | No |
| 6 | TGIF | TGIF Entrez, Source | TGFB-induced factor (TALE family homeobox) | 1094 | 0.137 | 0.1183 | No |
| 7 | SLC2A1 | SLC2A1 Entrez, Source | solute carrier family 2 (facilitated glucose transporter), member 1 | 1110 | 0.135 | 0.1339 | No |
| 8 | PLAG1 | PLAG1 Entrez, Source | pleiomorphic adenoma gene 1 | 1816 | 0.092 | 0.0920 | No |
| 9 | WNT5B | WNT5B Entrez, Source | wingless-type MMTV integration site family, member 5B | 2142 | 0.079 | 0.0773 | No |
| 10 | BCL2L1 | BCL2L1 Entrez, Source | BCL2-like 1 | 3166 | 0.050 | 0.0062 | No |
| 11 | RAP1B | RAP1B Entrez, Source | RAP1B, member of RAS oncogene family | 3177 | 0.050 | 0.0117 | No |
| 12 | ELOVL6 | ELOVL6 Entrez, Source | ELOVL family member 6, elongation of long chain fatty acids (FEN1/Elo2, SUR4/Elo3-like, yeast) | 3278 | 0.048 | 0.0100 | No |
| 13 | HERC4 | HERC4 Entrez, Source | hect domain and RLD 4 | 3315 | 0.047 | 0.0131 | No |
| 14 | PJA2 | PJA2 Entrez, Source | praja 2, RING-H2 motif containing | 3492 | 0.043 | 0.0051 | No |
| 15 | FZD1 | FZD1 Entrez, Source | frizzled homolog 1 (Drosophila) | 3493 | 0.043 | 0.0104 | No |
| 16 | NCOA6 | NCOA6 Entrez, Source | nuclear receptor coactivator 6 | 3527 | 0.042 | 0.0132 | No |
| 17 | GTF2B | GTF2B Entrez, Source | general transcription factor IIB | 4065 | 0.032 | -0.0235 | No |
| 18 | RAB5A | RAB5A Entrez, Source | RAB5A, member RAS oncogene family | 4218 | 0.029 | -0.0314 | No |
| 19 | POLD3 | POLD3 Entrez, Source | polymerase (DNA-directed), delta 3, accessory subunit | 4225 | 0.029 | -0.0283 | No |
| 20 | CLCN7 | CLCN7 Entrez, Source | chloride channel 7 | 4535 | 0.023 | -0.0488 | No |
| 21 | STK17B | STK17B Entrez, Source | serine/threonine kinase 17b (apoptosis-inducing) | 4570 | 0.022 | -0.0487 | No |
| 22 | FNTA | FNTA Entrez, Source | farnesyltransferase, CAAX box, alpha | 4587 | 0.022 | -0.0472 | No |
| 23 | RAB1A | RAB1A Entrez, Source | RAB1A, member RAS oncogene family | 4611 | 0.021 | -0.0463 | No |
| 24 | SON | SON Entrez, Source | SON DNA binding protein | 5330 | 0.010 | -0.0993 | No |
| 25 | CDC5L | CDC5L Entrez, Source | CDC5 cell division cycle 5-like (S. pombe) | 5412 | 0.009 | -0.1044 | No |
| 26 | MAPK14 | MAPK14 Entrez, Source | mitogen-activated protein kinase 14 | 5554 | 0.007 | -0.1142 | No |
| 27 | NBN | NBN Entrez, Source | nibrin | 5580 | 0.006 | -0.1153 | No |
| 28 | CDK7 | CDK7 Entrez, Source | cyclin-dependent kinase 7 (MO15 homolog, Xenopus laevis, cdk-activating kinase) | 5591 | 0.006 | -0.1154 | No |
| 29 | CDC42BPA | CDC42BPA Entrez, Source | CDC42 binding protein kinase alpha (DMPK-like) | 5723 | 0.004 | -0.1248 | No |
| 30 | TOP1 | TOP1 Entrez, Source | topoisomerase (DNA) I | 6078 | -0.001 | -0.1513 | No |
| 31 | PSMB1 | PSMB1 Entrez, Source | proteasome (prosome, macropain) subunit, beta type, 1 | 6487 | -0.007 | -0.1812 | No |
| 32 | HSPA8 | HSPA8 Entrez, Source | heat shock 70kDa protein 8 | 6613 | -0.009 | -0.1895 | No |
| 33 | BIRC2 | BIRC2 Entrez, Source | baculoviral IAP repeat-containing 2 | 6621 | -0.009 | -0.1889 | No |
| 34 | CXXC5 | CXXC5 Entrez, Source | CXXC finger 5 | 6698 | -0.011 | -0.1933 | No |
| 35 | MCTS1 | MCTS1 Entrez, Source | malignant T cell amplified sequence 1 | 6712 | -0.011 | -0.1929 | No |
| 36 | ERCC3 | ERCC3 Entrez, Source | excision repair cross-complementing rodent repair deficiency, complementation group 3 (xeroderma pigmentosum group B complementing) | 7085 | -0.016 | -0.2190 | No |
| 37 | PPP2R1B | PPP2R1B Entrez, Source | protein phosphatase 2 (formerly 2A), regulatory subunit A (PR 65), beta isoform | 7092 | -0.017 | -0.2174 | No |
| 38 | CDH11 | CDH11 Entrez, Source | cadherin 11, type 2, OB-cadherin (osteoblast) | 7101 | -0.017 | -0.2159 | No |
| 39 | SIRPA | SIRPA Entrez, Source | signal-regulatory protein alpha | 7460 | -0.022 | -0.2402 | No |
| 40 | SFRS9 | SFRS9 Entrez, Source | splicing factor, arginine/serine-rich 9 | 7495 | -0.022 | -0.2400 | No |
| 41 | PPP1CC | PPP1CC Entrez, Source | protein phosphatase 1, catalytic subunit, gamma isoform | 7701 | -0.026 | -0.2523 | No |
| 42 | DNAJA2 | DNAJA2 Entrez, Source | DnaJ (Hsp40) homolog, subfamily A, member 2 | 8107 | -0.032 | -0.2789 | No |
| 43 | PSMC2 | PSMC2 Entrez, Source | proteasome (prosome, macropain) 26S subunit, ATPase, 2 | 8346 | -0.037 | -0.2924 | No |
| 44 | THRAP3 | THRAP3 Entrez, Source | thyroid hormone receptor associated protein 3 | 8530 | -0.039 | -0.3013 | No |
| 45 | CCNA2 | CCNA2 Entrez, Source | cyclin A2 | 8927 | -0.046 | -0.3256 | No |
| 46 | CSNK2A1 | CSNK2A1 Entrez, Source | casein kinase 2, alpha 1 polypeptide | 8931 | -0.046 | -0.3201 | No |
| 47 | UBE2D3 | UBE2D3 Entrez, Source | ubiquitin-conjugating enzyme E2D 3 (UBC4/5 homolog, yeast) | 8991 | -0.047 | -0.3188 | No |
| 48 | PSMA4 | PSMA4 Entrez, Source | proteasome (prosome, macropain) subunit, alpha type, 4 | 9027 | -0.048 | -0.3155 | No |
| 49 | EIF3S10 | EIF3S10 Entrez, Source | eukaryotic translation initiation factor 3, subunit 10 theta, 150/170kDa | 9044 | -0.048 | -0.3108 | No |
| 50 | UBE2D2 | UBE2D2 Entrez, Source | ubiquitin-conjugating enzyme E2D 2 (UBC4/5 homolog, yeast) | 9056 | -0.048 | -0.3057 | No |
| 51 | RAB2 | RAB2 Entrez, Source | RAB2, member RAS oncogene family | 9159 | -0.050 | -0.3073 | No |
| 52 | TMPO | TMPO Entrez, Source | thymopoietin | 9189 | -0.050 | -0.3033 | No |
| 53 | CDC23 | CDC23 Entrez, Source | CDC23 (cell division cycle 23, yeast, homolog) | 9367 | -0.054 | -0.3100 | No |
| 54 | CRLF3 | CRLF3 Entrez, Source | cytokine receptor-like factor 3 | 9382 | -0.054 | -0.3044 | No |
| 55 | HSPE1 | HSPE1 Entrez, Source | heat shock 10kDa protein 1 (chaperonin 10) | 9837 | -0.063 | -0.3309 | No |
| 56 | PLK1 | PLK1 Entrez, Source | polo-like kinase 1 (Drosophila) | 10056 | -0.067 | -0.3391 | No |
| 57 | JMJD1C | JMJD1C Entrez, Source | jumonji domain containing 1C | 10296 | -0.072 | -0.3482 | Yes |
| 58 | MID1IP1 | MID1IP1 Entrez, Source | MID1 interacting protein 1 (gastrulation specific G12 homolog (zebrafish)) | 10305 | -0.073 | -0.3398 | Yes |
| 59 | BUB3 | BUB3 Entrez, Source | BUB3 budding uninhibited by benzimidazoles 3 homolog (yeast) | 10355 | -0.074 | -0.3345 | Yes |
| 60 | ATF1 | ATF1 Entrez, Source | activating transcription factor 1 | 10486 | -0.077 | -0.3347 | Yes |
| 61 | RABGGTB | RABGGTB Entrez, Source | Rab geranylgeranyltransferase, beta subunit | 10595 | -0.080 | -0.3331 | Yes |
| 62 | PSMC6 | PSMC6 Entrez, Source | proteasome (prosome, macropain) 26S subunit, ATPase, 6 | 10626 | -0.080 | -0.3254 | Yes |
| 63 | ARFGAP1 | ARFGAP1 Entrez, Source | ADP-ribosylation factor GTPase activating protein 1 | 10668 | -0.082 | -0.3185 | Yes |
| 64 | PSMA5 | PSMA5 Entrez, Source | proteasome (prosome, macropain) subunit, alpha type, 5 | 10870 | -0.087 | -0.3229 | Yes |
| 65 | CCNH | CCNH Entrez, Source | cyclin H | 10928 | -0.089 | -0.3162 | Yes |
| 66 | FBXL3 | FBXL3 Entrez, Source | F-box and leucine-rich repeat protein 3 | 10974 | -0.090 | -0.3086 | Yes |
| 67 | PRIM2A | PRIM2A Entrez, Source | primase, polypeptide 2A, 58kDa | 10985 | -0.090 | -0.2983 | Yes |
| 68 | SUPV3L1 | SUPV3L1 Entrez, Source | suppressor of var1, 3-like 1 (S. cerevisiae) | 11025 | -0.091 | -0.2900 | Yes |
| 69 | TOP2A | TOP2A Entrez, Source | topoisomerase (DNA) II alpha 170kDa | 11066 | -0.092 | -0.2816 | Yes |
| 70 | SNIP1 | SNIP1 Entrez, Source | Smad nuclear interacting protein 1 | 11119 | -0.094 | -0.2740 | Yes |
| 71 | PCNA | PCNA Entrez, Source | proliferating cell nuclear antigen | 11131 | -0.094 | -0.2633 | Yes |
| 72 | HDAC2 | HDAC2 Entrez, Source | histone deacetylase 2 | 11182 | -0.095 | -0.2553 | Yes |
| 73 | HSPH1 | HSPH1 Entrez, Source | heat shock 105kDa/110kDa protein 1 | 11297 | -0.099 | -0.2518 | Yes |
| 74 | CRSP6 | CRSP6 Entrez, Source | cofactor required for Sp1 transcriptional activation, subunit 6, 77kDa | 11432 | -0.103 | -0.2491 | Yes |
| 75 | CCT4 | CCT4 Entrez, Source | chaperonin containing TCP1, subunit 4 (delta) | 11590 | -0.109 | -0.2476 | Yes |
| 76 | NMI | NMI Entrez, Source | N-myc (and STAT) interactor | 11644 | -0.111 | -0.2379 | Yes |
| 77 | RBM17 | RBM17 Entrez, Source | RNA binding motif protein 17 | 11745 | -0.115 | -0.2313 | Yes |
| 78 | OGT | OGT Entrez, Source | O-linked N-acetylglucosamine (GlcNAc) transferase (UDP-N-acetylglucosamine:polypeptide-N-acetylglucosaminyl transferase) | 11749 | -0.115 | -0.2174 | Yes |
| 79 | PRKCBP1 | PRKCBP1 Entrez, Source | protein kinase C binding protein 1 | 11848 | -0.119 | -0.2101 | Yes |
| 80 | FOSL1 | FOSL1 Entrez, Source | FOS-like antigen 1 | 11938 | -0.123 | -0.2017 | Yes |
| 81 | PANX1 | PANX1 Entrez, Source | pannexin 1 | 12001 | -0.127 | -0.1907 | Yes |
| 82 | XBP1 | XBP1 Entrez, Source | X-box binding protein 1 | 12147 | -0.134 | -0.1852 | Yes |
| 83 | TARDBP | TARDBP Entrez, Source | TAR DNA binding protein | 12292 | -0.142 | -0.1786 | Yes |
| 84 | NUFIP1 | NUFIP1 Entrez, Source | nuclear fragile X mental retardation protein interacting protein 1 | 12295 | -0.142 | -0.1612 | Yes |
| 85 | LBR | LBR Entrez, Source | lamin B receptor | 12381 | -0.148 | -0.1495 | Yes |
| 86 | DDX20 | DDX20 Entrez, Source | DEAD (Asp-Glu-Ala-Asp) box polypeptide 20 | 12437 | -0.150 | -0.1351 | Yes |
| 87 | PSME3 | PSME3 Entrez, Source | proteasome (prosome, macropain) activator subunit 3 (PA28 gamma; Ki) | 12468 | -0.152 | -0.1186 | Yes |
| 88 | CDC27 | CDC27 Entrez, Source | cell division cycle 27 | 12571 | -0.160 | -0.1066 | Yes |
| 89 | USP3 | USP3 Entrez, Source | ubiquitin specific peptidase 3 | 12588 | -0.162 | -0.0879 | Yes |
| 90 | MTIF2 | MTIF2 Entrez, Source | mitochondrial translational initiation factor 2 | 12601 | -0.163 | -0.0686 | Yes |
| 91 | TXNRD1 | TXNRD1 Entrez, Source | thioredoxin reductase 1 | 12775 | -0.181 | -0.0594 | Yes |
| 92 | TDG | TDG Entrez, Source | thymine-DNA glycosylase | 12893 | -0.199 | -0.0438 | Yes |
| 93 | MAT2A | MAT2A Entrez, Source | methionine adenosyltransferase II, alpha | 13326 | -0.630 | 0.0012 | Yes |