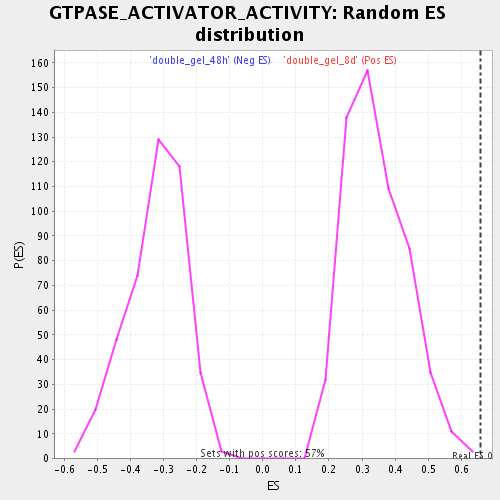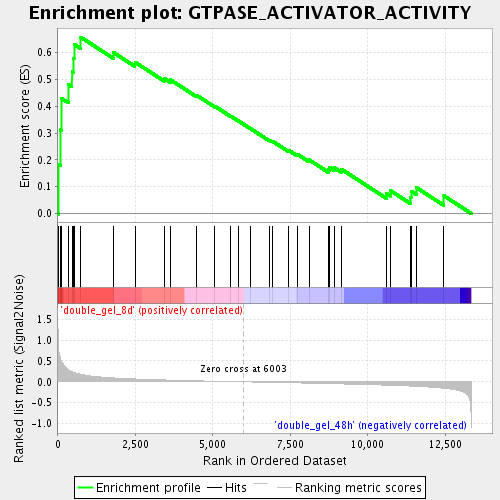
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | double_gel_and_single_gel_rma_expression_values_collapsed_to_symbols.class.cls #double_gel_8d_versus_double_gel_48h.class.cls #double_gel_8d_versus_double_gel_48h_repos |
| Phenotype | class.cls#double_gel_8d_versus_double_gel_48h_repos |
| Upregulated in class | double_gel_8d |
| GeneSet | GTPASE_ACTIVATOR_ACTIVITY |
| Enrichment Score (ES) | 0.657145 |
| Normalized Enrichment Score (NES) | 1.921541 |
| Nominal p-value | 0.001754386 |
| FDR q-value | 0.010059115 |
| FWER p-Value | 0.506 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | RGS2 | RGS2 Entrez, Source | regulator of G-protein signalling 2, 24kDa | 25 | 0.771 | 0.1814 | Yes |
| 2 | RGS1 | RGS1 Entrez, Source | regulator of G-protein signalling 1 | 75 | 0.565 | 0.3120 | Yes |
| 3 | RGS16 | RGS16 Entrez, Source | regulator of G-protein signalling 16 | 103 | 0.505 | 0.4301 | Yes |
| 4 | RGS3 | RGS3 Entrez, Source | regulator of G-protein signalling 3 | 338 | 0.287 | 0.4807 | Yes |
| 5 | RGS4 | RGS4 Entrez, Source | regulator of G-protein signalling 4 | 454 | 0.244 | 0.5300 | Yes |
| 6 | RGS5 | RGS5 Entrez, Source | regulator of G-protein signalling 5 | 513 | 0.224 | 0.5788 | Yes |
| 7 | CDC42EP2 | CDC42EP2 Entrez, Source | CDC42 effector protein (Rho GTPase binding) 2 | 522 | 0.222 | 0.6310 | Yes |
| 8 | ALDH1A1 | ALDH1A1 Entrez, Source | aldehyde dehydrogenase 1 family, member A1 | 739 | 0.178 | 0.6571 | Yes |
| 9 | RALBP1 | RALBP1 Entrez, Source | ralA binding protein 1 | 1792 | 0.093 | 0.6003 | No |
| 10 | ERRFI1 | ERRFI1 Entrez, Source | ERBB receptor feedback inhibitor 1 | 2488 | 0.068 | 0.5643 | No |
| 11 | CENTA1 | CENTA1 Entrez, Source | centaurin, alpha 1 | 3441 | 0.044 | 0.5033 | No |
| 12 | RAP1GAP | RAP1GAP Entrez, Source | RAP1 GTPase activating protein | 3624 | 0.040 | 0.4992 | No |
| 13 | MYO9B | MYO9B Entrez, Source | myosin IXB | 4486 | 0.024 | 0.4402 | No |
| 14 | RASA3 | RASA3 Entrez, Source | RAS p21 protein activator 3 | 5066 | 0.014 | 0.4000 | No |
| 15 | ARHGAP4 | ARHGAP4 Entrez, Source | Rho GTPase activating protein 4 | 5575 | 0.006 | 0.3633 | No |
| 16 | CHN2 | CHN2 Entrez, Source | chimerin (chimaerin) 2 | 5842 | 0.002 | 0.3439 | No |
| 17 | RASA2 | RASA2 Entrez, Source | RAS p21 protein activator 2 | 6217 | -0.003 | 0.3166 | No |
| 18 | RGS6 | RGS6 Entrez, Source | regulator of G-protein signalling 6 | 6832 | -0.013 | 0.2734 | No |
| 19 | RGS12 | RGS12 Entrez, Source | regulator of G-protein signalling 12 | 6916 | -0.014 | 0.2705 | No |
| 20 | RGS9 | RGS9 Entrez, Source | regulator of G-protein signalling 9 | 7456 | -0.022 | 0.2352 | No |
| 21 | CENTD2 | CENTD2 Entrez, Source | centaurin, delta 2 | 7725 | -0.026 | 0.2213 | No |
| 22 | ARHGAP5 | ARHGAP5 Entrez, Source | Rho GTPase activating protein 5 | 8103 | -0.032 | 0.2007 | No |
| 23 | ARHGAP27 | ARHGAP27 Entrez, Source | Rho GTPase activating protein 27 | 8729 | -0.043 | 0.1639 | No |
| 24 | RAB3GAP2 | RAB3GAP2 Entrez, Source | RAB3 GTPase activating protein subunit 2 (non-catalytic) | 8768 | -0.043 | 0.1713 | No |
| 25 | TSC2 | TSC2 Entrez, Source | tuberous sclerosis 2 | 8912 | -0.046 | 0.1714 | No |
| 26 | DLC1 | DLC1 Entrez, Source | deleted in liver cancer 1 | 9141 | -0.050 | 0.1661 | No |
| 27 | RANGAP1 | RANGAP1 Entrez, Source | Ran GTPase activating protein 1 | 10603 | -0.080 | 0.0753 | No |
| 28 | NF1 | NF1 Entrez, Source | neurofibromin 1 (neurofibromatosis, von Recklinghausen disease, Watson disease) | 10739 | -0.084 | 0.0851 | No |
| 29 | SIPA1 | SIPA1 Entrez, Source | signal-induced proliferation-associated gene 1 | 11380 | -0.101 | 0.0611 | No |
| 30 | THY1 | THY1 Entrez, Source | Thy-1 cell surface antigen | 11400 | -0.102 | 0.0839 | No |
| 31 | RGS14 | RGS14 Entrez, Source | regulator of G-protein signalling 14 | 11575 | -0.108 | 0.0966 | No |
| 32 | RASA1 | RASA1 Entrez, Source | RAS p21 protein activator (GTPase activating protein) 1 | 12457 | -0.152 | 0.0665 | No |