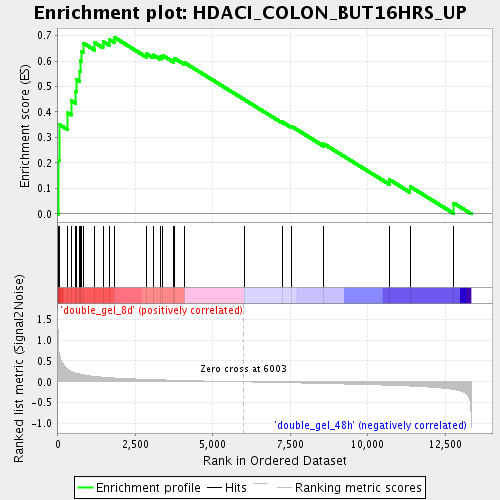
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | double_gel_and_single_gel_rma_expression_values_collapsed_to_symbols.class.cls #double_gel_8d_versus_double_gel_48h.class.cls #double_gel_8d_versus_double_gel_48h_repos |
| Phenotype | class.cls#double_gel_8d_versus_double_gel_48h_repos |
| Upregulated in class | double_gel_8d |
| GeneSet | HDACI_COLON_BUT16HRS_UP |
| Enrichment Score (ES) | 0.6931975 |
| Normalized Enrichment Score (NES) | 2.0075421 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.004178716 |
| FWER p-Value | 0.18 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | FOS | FOS Entrez, Source | v-fos FBJ murine osteosarcoma viral oncogene homolog | 9 | 0.936 | 0.2113 | Yes |
| 2 | SERPINE1 | SERPINE1 Entrez, Source | serpin peptidase inhibitor, clade E (nexin, plasminogen activator inhibitor type 1), member 1 | 56 | 0.628 | 0.3499 | Yes |
| 3 | ZSWIM6 | ZSWIM6 Entrez, Source | zinc finger, SWIM-type containing 6 | 320 | 0.292 | 0.3963 | Yes |
| 4 | RHBDF1 | RHBDF1 Entrez, Source | rhomboid 5 homolog 1 (Drosophila) | 431 | 0.250 | 0.4446 | Yes |
| 5 | IGFBP3 | IGFBP3 Entrez, Source | insulin-like growth factor binding protein 3 | 571 | 0.212 | 0.4821 | Yes |
| 6 | PTPN1 | PTPN1 Entrez, Source | protein tyrosine phosphatase, non-receptor type 1 | 599 | 0.207 | 0.5269 | Yes |
| 7 | RHOB | RHOB Entrez, Source | ras homolog gene family, member B | 709 | 0.184 | 0.5603 | Yes |
| 8 | GABBR1 | GABBR1 Entrez, Source | gamma-aminobutyric acid (GABA) B receptor, 1 | 722 | 0.182 | 0.6006 | Yes |
| 9 | CALCOCO1 | CALCOCO1 Entrez, Source | calcium binding and coiled-coil domain 1 | 771 | 0.174 | 0.6364 | Yes |
| 10 | NEU1 | NEU1 Entrez, Source | sialidase 1 (lysosomal sialidase) | 827 | 0.166 | 0.6698 | Yes |
| 11 | ANXA5 | ANXA5 Entrez, Source | annexin A5 | 1191 | 0.129 | 0.6717 | Yes |
| 12 | IL17RE | IL17RE Entrez, Source | interleukin 17 receptor E | 1457 | 0.110 | 0.6767 | Yes |
| 13 | BIRC4 | BIRC4 Entrez, Source | baculoviral IAP repeat-containing 4 | 1664 | 0.099 | 0.6837 | Yes |
| 14 | PLAG1 | PLAG1 Entrez, Source | pleiomorphic adenoma gene 1 | 1816 | 0.092 | 0.6932 | Yes |
| 15 | MAP1LC3B | MAP1LC3B Entrez, Source | microtubule-associated protein 1 light chain 3 beta | 2854 | 0.059 | 0.6286 | No |
| 16 | INPP5E | INPP5E Entrez, Source | inositol polyphosphate-5-phosphatase, 72 kDa | 3078 | 0.052 | 0.6237 | No |
| 17 | CDH1 | CDH1 Entrez, Source | cadherin 1, type 1, E-cadherin (epithelial) | 3296 | 0.047 | 0.6181 | No |
| 18 | ROCK1 | ROCK1 Entrez, Source | Rho-associated, coiled-coil containing protein kinase 1 | 3389 | 0.045 | 0.6215 | No |
| 19 | CLU | CLU Entrez, Source | clusterin | 3739 | 0.038 | 0.6039 | No |
| 20 | SPARCL1 | SPARCL1 Entrez, Source | SPARC-like 1 (mast9, hevin) | 3771 | 0.037 | 0.6100 | No |
| 21 | CAP1 | CAP1 Entrez, Source | CAP, adenylate cyclase-associated protein 1 (yeast) | 4084 | 0.031 | 0.5936 | No |
| 22 | CRYL1 | CRYL1 Entrez, Source | crystallin, lambda 1 | 6010 | -0.000 | 0.4490 | No |
| 23 | DNM3 | DNM3 Entrez, Source | dynamin 3 | 7241 | -0.019 | 0.3609 | No |
| 24 | GNL1 | GNL1 Entrez, Source | guanine nucleotide binding protein-like 1 | 7550 | -0.023 | 0.3431 | No |
| 25 | FLNB | FLNB Entrez, Source | filamin B, beta (actin binding protein 278) | 8583 | -0.040 | 0.2746 | No |
| 26 | PPM2C | PPM2C Entrez, Source | protein phosphatase 2C, magnesium-dependent, catalytic subunit | 10699 | -0.083 | 0.1345 | No |
| 27 | TSNAX | TSNAX Entrez, Source | translin-associated factor X | 11363 | -0.101 | 0.1075 | No |
| 28 | ID2 | ID2 Entrez, Source | inhibitor of DNA binding 2, dominant negative helix-loop-helix protein | 12777 | -0.181 | 0.0424 | No |

