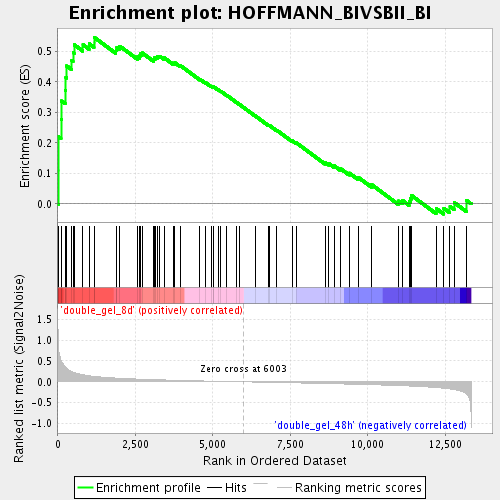
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | double_gel_and_single_gel_rma_expression_values_collapsed_to_symbols.class.cls #double_gel_8d_versus_double_gel_48h.class.cls #double_gel_8d_versus_double_gel_48h_repos |
| Phenotype | class.cls#double_gel_8d_versus_double_gel_48h_repos |
| Upregulated in class | double_gel_8d |
| GeneSet | HOFFMANN_BIVSBII_BI |
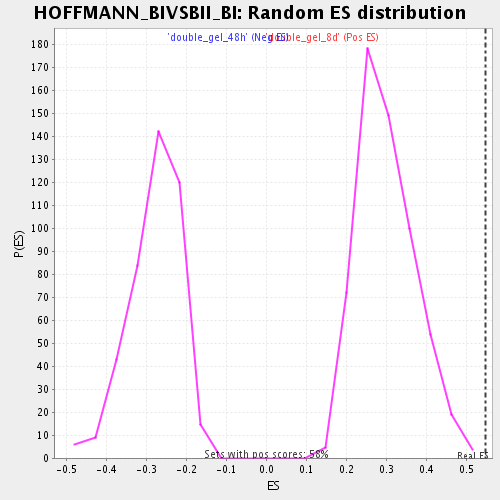
| Enrichment Score (ES) | 0.5464856 |
| Normalized Enrichment Score (NES) | 1.8177606 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.022840457 |
| FWER p-Value | 0.933 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | CCND2 | CCND2 Entrez, Source | cyclin D2 | 10 | 0.885 | 0.1103 | Yes |
| 2 | ANXA1 | ANXA1 Entrez, Source | annexin A1 | 11 | 0.884 | 0.2211 | Yes |
| 3 | EMP1 | EMP1 Entrez, Source | epithelial membrane protein 1 | 101 | 0.510 | 0.2784 | Yes |
| 4 | TNFAIP6 | TNFAIP6 Entrez, Source | tumor necrosis factor, alpha-induced protein 6 | 116 | 0.483 | 0.3379 | Yes |
| 5 | FXYD5 | FXYD5 Entrez, Source | FXYD domain containing ion transport regulator 5 | 237 | 0.355 | 0.3734 | Yes |
| 6 | AHNAK | AHNAK Entrez, Source | AHNAK nucleoprotein (desmoyokin) | 255 | 0.338 | 0.4144 | Yes |
| 7 | CD9 | CD9 Entrez, Source | CD9 molecule | 283 | 0.320 | 0.4526 | Yes |
| 8 | PKIB | PKIB Entrez, Source | protein kinase (cAMP-dependent, catalytic) inhibitor beta | 446 | 0.246 | 0.4712 | Yes |
| 9 | ITM2C | ITM2C Entrez, Source | integral membrane protein 2C | 491 | 0.231 | 0.4969 | Yes |
| 10 | AKR1C6 | 527 | 0.221 | 0.5220 | Yes | ||
| 11 | LMO4 | LMO4 Entrez, Source | LIM domain only 4 | 808 | 0.169 | 0.5221 | Yes |
| 12 | HEXA | HEXA Entrez, Source | hexosaminidase A (alpha polypeptide) | 1007 | 0.146 | 0.5255 | Yes |
| 13 | TAX1BP3 | TAX1BP3 Entrez, Source | Tax1 (human T-cell leukemia virus type I) binding protein 3 | 1166 | 0.131 | 0.5300 | Yes |
| 14 | CHRNB1 | CHRNB1 Entrez, Source | cholinergic receptor, nicotinic, beta 1 (muscle) | 1167 | 0.131 | 0.5465 | Yes |
| 15 | RUNX2 | RUNX2 Entrez, Source | runt-related transcription factor 2 | 1874 | 0.090 | 0.5046 | No |
| 16 | PIK4CB | PIK4CB Entrez, Source | phosphatidylinositol 4-kinase, catalytic, beta polypeptide | 1901 | 0.089 | 0.5137 | No |
| 17 | TCN2 | TCN2 Entrez, Source | transcobalamin II; macrocytic anemia | 1991 | 0.085 | 0.5177 | No |
| 18 | TLOC1 | TLOC1 Entrez, Source | translocation protein 1 | 2571 | 0.066 | 0.4823 | No |
| 19 | MAPT | MAPT Entrez, Source | microtubule-associated protein tau | 2638 | 0.064 | 0.4854 | No |
| 20 | ZYX | ZYX Entrez, Source | zyxin | 2654 | 0.063 | 0.4922 | No |
| 21 | EMB | EMB Entrez, Source | embigin homolog (mouse) | 2717 | 0.062 | 0.4953 | No |
| 22 | SQLE | SQLE Entrez, Source | squalene epoxidase | 3091 | 0.052 | 0.4738 | No |
| 23 | ADA | ADA Entrez, Source | adenosine deaminase | 3106 | 0.052 | 0.4792 | No |
| 24 | MAGED2 | MAGED2 Entrez, Source | melanoma antigen family D, 2 | 3164 | 0.050 | 0.4812 | No |
| 25 | ACTN1 | ACTN1 Entrez, Source | actinin, alpha 1 | 3213 | 0.049 | 0.4838 | No |
| 26 | ELOVL6 | ELOVL6 Entrez, Source | ELOVL family member 6, elongation of long chain fatty acids (FEN1/Elo2, SUR4/Elo3-like, yeast) | 3278 | 0.048 | 0.4850 | No |
| 27 | SOCS2 | SOCS2 Entrez, Source | suppressor of cytokine signaling 2 | 3428 | 0.044 | 0.4793 | No |
| 28 | HDAC5 | HDAC5 Entrez, Source | histone deacetylase 5 | 3742 | 0.038 | 0.4605 | No |
| 29 | CACNB3 | CACNB3 Entrez, Source | calcium channel, voltage-dependent, beta 3 subunit | 3774 | 0.037 | 0.4628 | No |
| 30 | ANXA2 | ANXA2 Entrez, Source | annexin A2 | 3949 | 0.034 | 0.4539 | No |
| 31 | HEMGN | HEMGN Entrez, Source | hemogen | 4584 | 0.022 | 0.4089 | No |
| 32 | SOCS1 | SOCS1 Entrez, Source | suppressor of cytokine signaling 1 | 4762 | 0.019 | 0.3980 | No |
| 33 | CRYAA | CRYAA Entrez, Source | crystallin, alpha A | 4950 | 0.016 | 0.3859 | No |
| 34 | PPP1CA | PPP1CA Entrez, Source | protein phosphatase 1, catalytic subunit, alpha isoform | 5008 | 0.015 | 0.3835 | No |
| 35 | IFIT2 | IFIT2 Entrez, Source | interferon-induced protein with tetratricopeptide repeats 2 | 5017 | 0.015 | 0.3848 | No |
| 36 | STAT4 | STAT4 Entrez, Source | signal transducer and activator of transcription 4 | 5174 | 0.012 | 0.3745 | No |
| 37 | GAP43 | GAP43 Entrez, Source | growth associated protein 43 | 5249 | 0.011 | 0.3703 | No |
| 38 | CTSL | CTSL Entrez, Source | cathepsin L | 5451 | 0.008 | 0.3562 | No |
| 39 | ENG | ENG Entrez, Source | endoglin (Osler-Rendu-Weber syndrome 1) | 5761 | 0.003 | 0.3333 | No |
| 40 | DAB2IP | DAB2IP Entrez, Source | DAB2 interacting protein | 5845 | 0.002 | 0.3274 | No |
| 41 | BCL2 | BCL2 Entrez, Source | B-cell CLL/lymphoma 2 | 6363 | -0.006 | 0.2891 | No |
| 42 | ATP6V0A1 | ATP6V0A1 Entrez, Source | ATPase, H+ transporting, lysosomal V0 subunit a1 | 6799 | -0.012 | 0.2579 | No |
| 43 | GRB7 | GRB7 Entrez, Source | growth factor receptor-bound protein 7 | 6828 | -0.013 | 0.2574 | No |
| 44 | CUBN | CUBN Entrez, Source | cubilin (intrinsic factor-cobalamin receptor) | 7059 | -0.016 | 0.2421 | No |
| 45 | RHOH | RHOH Entrez, Source | ras homolog gene family, member H | 7559 | -0.024 | 0.2075 | No |
| 46 | THRA | THRA Entrez, Source | thyroid hormone receptor, alpha (erythroblastic leukemia viral (v-erb-a) oncogene homolog, avian) | 7692 | -0.026 | 0.2007 | No |
| 47 | EDN2 | EDN2 Entrez, Source | endothelin 2 | 8626 | -0.041 | 0.1356 | No |
| 48 | ST3GAL6 | ST3GAL6 Entrez, Source | ST3 beta-galactoside alpha-2,3-sialyltransferase 6 | 8734 | -0.043 | 0.1329 | No |
| 49 | CCL5 | CCL5 Entrez, Source | chemokine (C-C motif) ligand 5 | 8913 | -0.046 | 0.1253 | No |
| 50 | GIMAP4 | GIMAP4 Entrez, Source | GTPase, IMAP family member 4 | 9106 | -0.049 | 0.1169 | No |
| 51 | MYEF2 | MYEF2 Entrez, Source | myelin expression factor 2 | 9401 | -0.054 | 0.1016 | No |
| 52 | TBXA2R | TBXA2R Entrez, Source | thromboxane A2 receptor | 9702 | -0.060 | 0.0865 | No |
| 53 | MAPK8IP3 | MAPK8IP3 Entrez, Source | mitogen-activated protein kinase 8 interacting protein 3 | 10124 | -0.069 | 0.0634 | No |
| 54 | PRIM2A | PRIM2A Entrez, Source | primase, polypeptide 2A, 58kDa | 10985 | -0.090 | 0.0100 | No |
| 55 | TACSTD2 | TACSTD2 Entrez, Source | tumor-associated calcium signal transducer 2 | 11118 | -0.094 | 0.0118 | No |
| 56 | ENPP1 | ENPP1 Entrez, Source | ectonucleotide pyrophosphatase/phosphodiesterase 1 | 11335 | -0.100 | 0.0081 | No |
| 57 | SIPA1 | SIPA1 Entrez, Source | signal-induced proliferation-associated gene 1 | 11380 | -0.101 | 0.0175 | No |
| 58 | THY1 | THY1 Entrez, Source | Thy-1 cell surface antigen | 11400 | -0.102 | 0.0288 | No |
| 59 | BIN1 | BIN1 Entrez, Source | bridging integrator 1 | 12208 | -0.137 | -0.0147 | No |
| 60 | KLF10 | KLF10 Entrez, Source | Kruppel-like factor 10 | 12453 | -0.151 | -0.0141 | No |
| 61 | PPIC | PPIC Entrez, Source | peptidylprolyl isomerase C (cyclophilin C) | 12653 | -0.168 | -0.0080 | No |
| 62 | MDN1 | MDN1 Entrez, Source | MDN1, midasin homolog (yeast) | 12790 | -0.183 | 0.0048 | No |
| 63 | LTB | LTB Entrez, Source | lymphotoxin beta (TNF superfamily, member 3) | 13186 | -0.293 | 0.0117 | No |