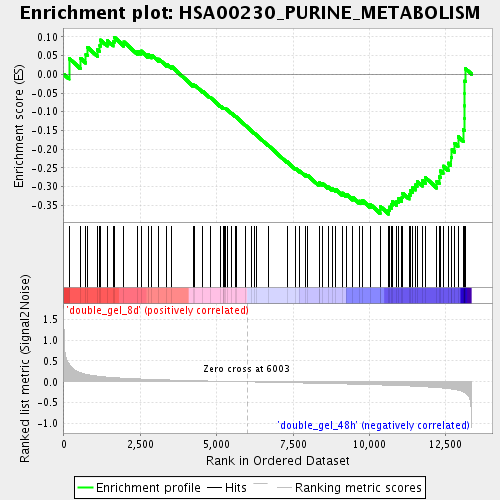
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | double_gel_and_single_gel_rma_expression_values_collapsed_to_symbols.class.cls #double_gel_8d_versus_double_gel_48h.class.cls #double_gel_8d_versus_double_gel_48h_repos |
| Phenotype | class.cls#double_gel_8d_versus_double_gel_48h_repos |
| Upregulated in class | double_gel_48h |
| GeneSet | HSA00230_PURINE_METABOLISM |
| Enrichment Score (ES) | -0.3734825 |
| Normalized Enrichment Score (NES) | -1.4103353 |
| Nominal p-value | 0.029411765 |
| FDR q-value | 0.33380204 |
| FWER p-Value | 1.0 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | AMPD3 | AMPD3 Entrez, Source | adenosine monophosphate deaminase (isoform E) | 167 | 0.426 | 0.0428 | No |
| 2 | GDA | GDA Entrez, Source | guanine deaminase | 537 | 0.219 | 0.0435 | No |
| 3 | PDE4C | PDE4C Entrez, Source | phosphodiesterase 4C, cAMP-specific (phosphodiesterase E1 dunce homolog, Drosophila) | 719 | 0.182 | 0.0535 | No |
| 4 | GUCY1B3 | GUCY1B3 Entrez, Source | guanylate cyclase 1, soluble, beta 3 | 766 | 0.175 | 0.0728 | No |
| 5 | GUCY2C | GUCY2C Entrez, Source | guanylate cyclase 2C (heat stable enterotoxin receptor) | 1098 | 0.136 | 0.0656 | No |
| 6 | PDE10A | PDE10A Entrez, Source | phosphodiesterase 10A | 1176 | 0.130 | 0.0767 | No |
| 7 | AMPD1 | AMPD1 Entrez, Source | adenosine monophosphate deaminase 1 (isoform M) | 1199 | 0.128 | 0.0917 | No |
| 8 | ENTPD2 | ENTPD2 Entrez, Source | ectonucleoside triphosphate diphosphohydrolase 2 | 1415 | 0.113 | 0.0901 | No |
| 9 | GUK1 | GUK1 Entrez, Source | guanylate kinase 1 | 1609 | 0.102 | 0.0889 | No |
| 10 | GMPR | GMPR Entrez, Source | guanosine monophosphate reductase | 1649 | 0.100 | 0.0989 | No |
| 11 | FHIT | FHIT Entrez, Source | fragile histidine triad gene | 1965 | 0.086 | 0.0864 | No |
| 12 | ADCY6 | ADCY6 Entrez, Source | adenylate cyclase 6 | 2414 | 0.070 | 0.0618 | No |
| 13 | NUDT2 | NUDT2 Entrez, Source | nudix (nucleoside diphosphate linked moiety X)-type motif 2 | 2523 | 0.067 | 0.0623 | No |
| 14 | ENTPD3 | ENTPD3 Entrez, Source | ectonucleoside triphosphate diphosphohydrolase 3 | 2753 | 0.061 | 0.0530 | No |
| 15 | ENTPD6 | ENTPD6 Entrez, Source | ectonucleoside triphosphate diphosphohydrolase 6 (putative function) | 2881 | 0.058 | 0.0510 | No |
| 16 | ADA | ADA Entrez, Source | adenosine deaminase | 3106 | 0.052 | 0.0408 | No |
| 17 | NME7 | NME7 Entrez, Source | non-metastatic cells 7, protein expressed in (nucleoside-diphosphate kinase) | 3369 | 0.046 | 0.0270 | No |
| 18 | PDE6H | PDE6H Entrez, Source | phosphodiesterase 6H, cGMP-specific, cone, gamma | 3524 | 0.042 | 0.0209 | No |
| 19 | POLD3 | POLD3 Entrez, Source | polymerase (DNA-directed), delta 3, accessory subunit | 4225 | 0.029 | -0.0281 | No |
| 20 | PDE11A | PDE11A Entrez, Source | phosphodiesterase 11A | 4284 | 0.028 | -0.0289 | No |
| 21 | AK1 | AK1 Entrez, Source | adenylate kinase 1 | 4543 | 0.022 | -0.0454 | No |
| 22 | ADCY5 | ADCY5 Entrez, Source | adenylate cyclase 5 | 4781 | 0.019 | -0.0609 | No |
| 23 | PDE8A | PDE8A Entrez, Source | phosphodiesterase 8A | 5133 | 0.013 | -0.0857 | No |
| 24 | GMPR2 | GMPR2 Entrez, Source | guanosine monophosphate reductase 2 | 5222 | 0.011 | -0.0909 | No |
| 25 | GUCY1A3 | GUCY1A3 Entrez, Source | guanylate cyclase 1, soluble, alpha 3 | 5232 | 0.011 | -0.0901 | No |
| 26 | PDE2A | PDE2A Entrez, Source | phosphodiesterase 2A, cGMP-stimulated | 5264 | 0.011 | -0.0910 | No |
| 27 | POLR2F | POLR2F Entrez, Source | polymerase (RNA) II (DNA directed) polypeptide F | 5278 | 0.010 | -0.0906 | No |
| 28 | PDE7A | PDE7A Entrez, Source | phosphodiesterase 7A | 5340 | 0.009 | -0.0940 | No |
| 29 | PDE1C | PDE1C Entrez, Source | phosphodiesterase 1C, calmodulin-dependent 70kDa | 5479 | 0.008 | -0.1034 | No |
| 30 | ADK | ADK Entrez, Source | adenosine kinase | 5620 | 0.005 | -0.1133 | No |
| 31 | ZNRD1 | ZNRD1 Entrez, Source | zinc ribbon domain containing 1 | 5635 | 0.005 | -0.1137 | No |
| 32 | POLR2G | POLR2G Entrez, Source | polymerase (RNA) II (DNA directed) polypeptide G | 5947 | 0.001 | -0.1370 | No |
| 33 | GUCY2D | GUCY2D Entrez, Source | guanylate cyclase 2D, membrane (retina-specific) | 6125 | -0.002 | -0.1501 | No |
| 34 | PDE4B | PDE4B Entrez, Source | phosphodiesterase 4B, cAMP-specific (phosphodiesterase E4 dunce homolog, Drosophila) | 6239 | -0.004 | -0.1582 | No |
| 35 | PDE4A | PDE4A Entrez, Source | phosphodiesterase 4A, cAMP-specific (phosphodiesterase E2 dunce homolog, Drosophila) | 6308 | -0.005 | -0.1627 | No |
| 36 | PDE3B | PDE3B Entrez, Source | phosphodiesterase 3B, cGMP-inhibited | 6687 | -0.011 | -0.1898 | No |
| 37 | POLR2C | POLR2C Entrez, Source | polymerase (RNA) II (DNA directed) polypeptide C, 33kDa | 7301 | -0.020 | -0.2335 | No |
| 38 | ADCY2 | ADCY2 Entrez, Source | adenylate cyclase 2 (brain) | 7570 | -0.024 | -0.2506 | No |
| 39 | NT5E | NT5E Entrez, Source | 5'-nucleotidase, ecto (CD73) | 7715 | -0.026 | -0.2581 | No |
| 40 | POLD4 | POLD4 Entrez, Source | polymerase (DNA-directed), delta 4 | 7902 | -0.029 | -0.2683 | No |
| 41 | PDE4D | PDE4D Entrez, Source | phosphodiesterase 4D, cAMP-specific (phosphodiesterase E3 dunce homolog, Drosophila) | 7982 | -0.030 | -0.2703 | No |
| 42 | ENTPD1 | ENTPD1 Entrez, Source | ectonucleoside triphosphate diphosphohydrolase 1 | 8353 | -0.037 | -0.2935 | No |
| 43 | CANT1 | CANT1 Entrez, Source | calcium activated nucleotidase 1 | 8366 | -0.037 | -0.2896 | No |
| 44 | PDE1A | PDE1A Entrez, Source | phosphodiesterase 1A, calmodulin-dependent | 8459 | -0.038 | -0.2915 | No |
| 45 | POLA2 | POLA2 Entrez, Source | polymerase (DNA directed), alpha 2 (70kD subunit) | 8655 | -0.041 | -0.3009 | No |
| 46 | HPRT1 | HPRT1 Entrez, Source | hypoxanthine phosphoribosyltransferase 1 (Lesch-Nyhan syndrome) | 8796 | -0.044 | -0.3058 | No |
| 47 | ADCY8 | ADCY8 Entrez, Source | adenylate cyclase 8 (brain) | 8892 | -0.045 | -0.3070 | No |
| 48 | AK3L1 | AK3L1 Entrez, Source | adenylate kinase 3-like 1 | 9110 | -0.049 | -0.3170 | No |
| 49 | PDE8B | PDE8B Entrez, Source | phosphodiesterase 8B | 9248 | -0.052 | -0.3206 | No |
| 50 | XDH | XDH Entrez, Source | xanthine dehydrogenase | 9455 | -0.055 | -0.3290 | No |
| 51 | GMPS | GMPS Entrez, Source | guanine monphosphate synthetase | 9684 | -0.060 | -0.3384 | No |
| 52 | NME2 | NME2 Entrez, Source | non-metastatic cells 2, protein (NM23B) expressed in | 9773 | -0.061 | -0.3371 | No |
| 53 | PDE9A | PDE9A Entrez, Source | phosphodiesterase 9A | 10033 | -0.067 | -0.3479 | No |
| 54 | GUCY1A2 | GUCY1A2 Entrez, Source | guanylate cyclase 1, soluble, alpha 2 | 10348 | -0.074 | -0.3620 | No |
| 55 | AK2 | AK2 Entrez, Source | adenylate kinase 2 | 10352 | -0.074 | -0.3527 | No |
| 56 | NPR2 | NPR2 Entrez, Source | natriuretic peptide receptor B/guanylate cyclase B (atrionatriuretic peptide receptor B) | 10629 | -0.081 | -0.3630 | Yes |
| 57 | ADCY3 | ADCY3 Entrez, Source | adenylate cyclase 3 | 10657 | -0.081 | -0.3545 | Yes |
| 58 | POLE3 | POLE3 Entrez, Source | polymerase (DNA directed), epsilon 3 (p17 subunit) | 10715 | -0.083 | -0.3479 | Yes |
| 59 | ATIC | ATIC Entrez, Source | 5-aminoimidazole-4-carboxamide ribonucleotide formyltransferase/IMP cyclohydrolase | 10742 | -0.084 | -0.3390 | Yes |
| 60 | NPR1 | NPR1 Entrez, Source | natriuretic peptide receptor A/guanylate cyclase A (atrionatriuretic peptide receptor A) | 10893 | -0.088 | -0.3389 | Yes |
| 61 | POLR1A | POLR1A Entrez, Source | polymerase (RNA) I polypeptide A, 194kDa | 10957 | -0.089 | -0.3320 | Yes |
| 62 | POLD1 | POLD1 Entrez, Source | polymerase (DNA directed), delta 1, catalytic subunit 125kDa | 11062 | -0.092 | -0.3279 | Yes |
| 63 | ENPP3 | ENPP3 Entrez, Source | ectonucleotide pyrophosphatase/phosphodiesterase 3 | 11082 | -0.093 | -0.3172 | Yes |
| 64 | NUDT9 | NUDT9 Entrez, Source | nudix (nucleoside diphosphate linked moiety X)-type motif 9 | 11295 | -0.098 | -0.3204 | Yes |
| 65 | ENPP1 | ENPP1 Entrez, Source | ectonucleotide pyrophosphatase/phosphodiesterase 1 | 11335 | -0.100 | -0.3104 | Yes |
| 66 | GART | GART Entrez, Source | phosphoribosylglycinamide formyltransferase, phosphoribosylglycinamide synthetase, phosphoribosylaminoimidazole synthetase | 11406 | -0.103 | -0.3023 | Yes |
| 67 | GUCY2F | GUCY2F Entrez, Source | guanylate cyclase 2F, retinal | 11499 | -0.106 | -0.2955 | Yes |
| 68 | PNPT1 | PNPT1 Entrez, Source | polyribonucleotide nucleotidyltransferase 1 | 11558 | -0.108 | -0.2858 | Yes |
| 69 | PAICS | PAICS Entrez, Source | phosphoribosylaminoimidazole carboxylase, phosphoribosylaminoimidazole succinocarboxamide synthetase | 11730 | -0.114 | -0.2838 | Yes |
| 70 | PRPS1 | PRPS1 Entrez, Source | phosphoribosyl pyrophosphate synthetase 1 | 11832 | -0.118 | -0.2761 | Yes |
| 71 | POLD2 | POLD2 Entrez, Source | polymerase (DNA directed), delta 2, regulatory subunit 50kDa | 12204 | -0.137 | -0.2863 | Yes |
| 72 | PRIM1 | PRIM1 Entrez, Source | primase, polypeptide 1, 49kDa | 12284 | -0.141 | -0.2738 | Yes |
| 73 | NME1 | NME1 Entrez, Source | non-metastatic cells 1, protein (NM23A) expressed in | 12324 | -0.144 | -0.2580 | Yes |
| 74 | NUDT5 | NUDT5 Entrez, Source | nudix (nucleoside diphosphate linked moiety X)-type motif 5 | 12409 | -0.149 | -0.2450 | Yes |
| 75 | ADCY4 | ADCY4 Entrez, Source | adenylate cyclase 4 | 12581 | -0.161 | -0.2370 | Yes |
| 76 | PDE5A | PDE5A Entrez, Source | phosphodiesterase 5A, cGMP-specific | 12668 | -0.170 | -0.2214 | Yes |
| 77 | PDE7B | PDE7B Entrez, Source | phosphodiesterase 7B | 12699 | -0.173 | -0.2011 | Yes |
| 78 | POLR1B | POLR1B Entrez, Source | polymerase (RNA) I polypeptide B, 128kDa | 12791 | -0.183 | -0.1841 | Yes |
| 79 | IMPDH2 | IMPDH2 Entrez, Source | IMP (inosine monophosphate) dehydrogenase 2 | 12899 | -0.199 | -0.1663 | Yes |
| 80 | NME6 | NME6 Entrez, Source | non-metastatic cells 6, protein expressed in (nucleoside-diphosphate kinase) | 13072 | -0.241 | -0.1479 | Yes |
| 81 | PPAT | PPAT Entrez, Source | phosphoribosyl pyrophosphate amidotransferase | 13102 | -0.253 | -0.1172 | Yes |
| 82 | PRPS2 | PRPS2 Entrez, Source | phosphoribosyl pyrophosphate synthetase 2 | 13109 | -0.256 | -0.0844 | Yes |
| 83 | PKLR | PKLR Entrez, Source | pyruvate kinase, liver and RBC | 13114 | -0.257 | -0.0513 | Yes |
| 84 | DCK | DCK Entrez, Source | deoxycytidine kinase | 13118 | -0.258 | -0.0179 | Yes |
| 85 | ENTPD5 | ENTPD5 Entrez, Source | ectonucleoside triphosphate diphosphohydrolase 5 | 13144 | -0.267 | 0.0149 | Yes |

