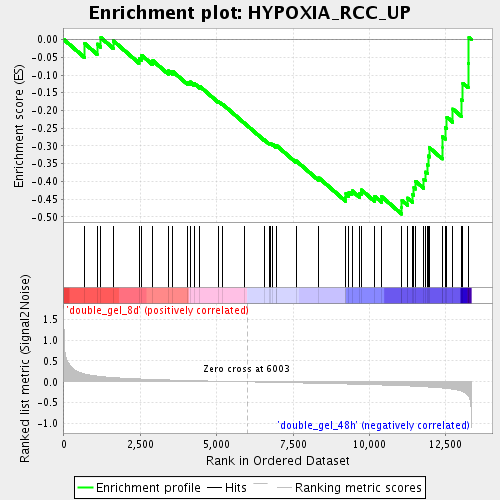
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | double_gel_and_single_gel_rma_expression_values_collapsed_to_symbols.class.cls #double_gel_8d_versus_double_gel_48h.class.cls #double_gel_8d_versus_double_gel_48h_repos |
| Phenotype | class.cls#double_gel_8d_versus_double_gel_48h_repos |
| Upregulated in class | double_gel_48h |
| GeneSet | HYPOXIA_RCC_UP |
| Enrichment Score (ES) | -0.49241126 |
| Normalized Enrichment Score (NES) | -1.7053274 |
| Nominal p-value | 0.006849315 |
| FDR q-value | 0.083486244 |
| FWER p-Value | 0.998 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | CD276 | CD276 Entrez, Source | CD276 molecule | 683 | 0.188 | -0.0114 | No |
| 2 | KIFC3 | KIFC3 Entrez, Source | kinesin family member C3 | 1087 | 0.137 | -0.0124 | No |
| 3 | GADD45B | GADD45B Entrez, Source | growth arrest and DNA-damage-inducible, beta | 1210 | 0.127 | 0.0054 | No |
| 4 | TES | TES Entrez, Source | testis derived transcript (3 LIM domains) | 1616 | 0.102 | -0.0034 | No |
| 5 | VIL2 | VIL2 Entrez, Source | villin 2 (ezrin) | 2459 | 0.069 | -0.0520 | No |
| 6 | MRPL41 | MRPL41 Entrez, Source | mitochondrial ribosomal protein L41 | 2546 | 0.066 | -0.0444 | No |
| 7 | DPM2 | DPM2 Entrez, Source | dolichyl-phosphate mannosyltransferase polypeptide 2, regulatory subunit | 2909 | 0.057 | -0.0595 | No |
| 8 | CEACAM1 | CEACAM1 Entrez, Source | carcinoembryonic antigen-related cell adhesion molecule 1 (biliary glycoprotein) | 3419 | 0.045 | -0.0882 | No |
| 9 | WDR6 | WDR6 Entrez, Source | WD repeat domain 6 | 3554 | 0.042 | -0.0895 | No |
| 10 | PECR | PECR Entrez, Source | peroxisomal trans-2-enoyl-CoA reductase | 4050 | 0.032 | -0.1199 | No |
| 11 | APLP2 | APLP2 Entrez, Source | amyloid beta (A4) precursor-like protein 2 | 4130 | 0.030 | -0.1194 | No |
| 12 | FHL1 | FHL1 Entrez, Source | four and a half LIM domains 1 | 4269 | 0.028 | -0.1238 | No |
| 13 | GADD45GIP1 | GADD45GIP1 Entrez, Source | growth arrest and DNA-damage-inducible, gamma interacting protein 1 | 4449 | 0.024 | -0.1321 | No |
| 14 | ARF5 | ARF5 Entrez, Source | ADP-ribosylation factor 5 | 5062 | 0.014 | -0.1752 | No |
| 15 | SSU72 | SSU72 Entrez, Source | SSU72 RNA polymerase II CTD phosphatase homolog (S. cerevisiae) | 5188 | 0.012 | -0.1820 | No |
| 16 | ATP5G3 | ATP5G3 Entrez, Source | ATP synthase, H+ transporting, mitochondrial F0 complex, subunit C3 (subunit 9) | 5923 | 0.001 | -0.2370 | No |
| 17 | YIPF5 | YIPF5 Entrez, Source | Yip1 domain family, member 5 | 6555 | -0.008 | -0.2827 | No |
| 18 | PDLIM7 | PDLIM7 Entrez, Source | PDZ and LIM domain 7 (enigma) | 6732 | -0.011 | -0.2936 | No |
| 19 | GTF2A2 | GTF2A2 Entrez, Source | general transcription factor IIA, 2, 12kDa | 6756 | -0.012 | -0.2928 | No |
| 20 | LRRC48 | LRRC48 Entrez, Source | leucine rich repeat containing 48 | 6825 | -0.013 | -0.2953 | No |
| 21 | RHOT2 | RHOT2 Entrez, Source | ras homolog gene family, member T2 | 6953 | -0.014 | -0.3018 | No |
| 22 | PRPF19 | PRPF19 Entrez, Source | PRP19/PSO4 pre-mRNA processing factor 19 homolog (S. cerevisiae) | 6964 | -0.014 | -0.2994 | No |
| 23 | HIRIP3 | HIRIP3 Entrez, Source | HIRA interacting protein 3 | 7603 | -0.024 | -0.3423 | No |
| 24 | TRSPAP1 | TRSPAP1 Entrez, Source | tRNA selenocysteine associated protein 1 | 8345 | -0.036 | -0.3903 | No |
| 25 | PPP2R5D | PPP2R5D Entrez, Source | protein phosphatase 2, regulatory subunit B (B56), delta isoform | 9222 | -0.051 | -0.4453 | No |
| 26 | RPL37 | RPL37 Entrez, Source | ribosomal protein L37 | 9228 | -0.051 | -0.4348 | No |
| 27 | MRPL13 | MRPL13 Entrez, Source | mitochondrial ribosomal protein L13 | 9328 | -0.053 | -0.4309 | No |
| 28 | CTSC | CTSC Entrez, Source | cathepsin C | 9429 | -0.055 | -0.4268 | No |
| 29 | GNG12 | GNG12 Entrez, Source | guanine nucleotide binding protein (G protein), gamma 12 | 9681 | -0.059 | -0.4330 | No |
| 30 | RPL3 | RPL3 Entrez, Source | ribosomal protein L3 | 9736 | -0.061 | -0.4242 | No |
| 31 | APEX1 | APEX1 Entrez, Source | APEX nuclease (multifunctional DNA repair enzyme) 1 | 10173 | -0.070 | -0.4421 | No |
| 32 | RAB14 | RAB14 Entrez, Source | RAB14, member RAS oncogene family | 10401 | -0.075 | -0.4433 | No |
| 33 | RPS15A | RPS15A Entrez, Source | ribosomal protein S15a | 11055 | -0.092 | -0.4728 | Yes |
| 34 | POLD1 | POLD1 Entrez, Source | polymerase (DNA directed), delta 1, catalytic subunit 125kDa | 11062 | -0.092 | -0.4537 | Yes |
| 35 | EWSR1 | EWSR1 Entrez, Source | Ewing sarcoma breakpoint region 1 | 11248 | -0.097 | -0.4469 | Yes |
| 36 | CAND1 | CAND1 Entrez, Source | cullin-associated and neddylation-dissociated 1 | 11414 | -0.103 | -0.4375 | Yes |
| 37 | METTL3 | METTL3 Entrez, Source | methyltransferase like 3 | 11454 | -0.104 | -0.4182 | Yes |
| 38 | CNTN5 | CNTN5 Entrez, Source | contactin 5 | 11504 | -0.106 | -0.3993 | Yes |
| 39 | SCYE1 | SCYE1 Entrez, Source | small inducible cytokine subfamily E, member 1 (endothelial monocyte-activating) | 11779 | -0.116 | -0.3952 | Yes |
| 40 | TMEM5 | TMEM5 Entrez, Source | transmembrane protein 5 | 11827 | -0.118 | -0.3736 | Yes |
| 41 | SERBP1 | SERBP1 Entrez, Source | SERPINE1 mRNA binding protein 1 | 11903 | -0.122 | -0.3534 | Yes |
| 42 | PPM1F | PPM1F Entrez, Source | protein phosphatase 1F (PP2C domain containing) | 11918 | -0.122 | -0.3283 | Yes |
| 43 | RPL22 | RPL22 Entrez, Source | ribosomal protein L22 | 11961 | -0.125 | -0.3050 | Yes |
| 44 | GMFB | GMFB Entrez, Source | glia maturation factor, beta | 12387 | -0.148 | -0.3055 | Yes |
| 45 | RPL39 | RPL39 Entrez, Source | ribosomal protein L39 | 12390 | -0.148 | -0.2741 | Yes |
| 46 | PARL | PARL Entrez, Source | presenilin associated, rhomboid-like | 12481 | -0.153 | -0.2483 | Yes |
| 47 | IDI1 | IDI1 Entrez, Source | isopentenyl-diphosphate delta isomerase 1 | 12532 | -0.157 | -0.2186 | Yes |
| 48 | SYNCRIP | SYNCRIP Entrez, Source | synaptotagmin binding, cytoplasmic RNA interacting protein | 12730 | -0.176 | -0.1959 | Yes |
| 49 | NOLC1 | NOLC1 Entrez, Source | nucleolar and coiled-body phosphoprotein 1 | 13015 | -0.221 | -0.1701 | Yes |
| 50 | SCARB2 | SCARB2 Entrez, Source | scavenger receptor class B, member 2 | 13055 | -0.235 | -0.1230 | Yes |
| 51 | KIF1B | KIF1B Entrez, Source | kinesin family member 1B | 13231 | -0.325 | -0.0669 | Yes |
| 52 | SLC3A2 | SLC3A2 Entrez, Source | solute carrier family 3 (activators of dibasic and neutral amino acid transport), member 2 | 13255 | -0.353 | 0.0065 | Yes |