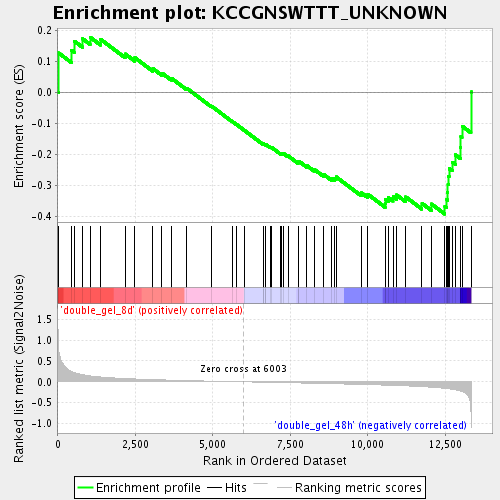
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | double_gel_and_single_gel_rma_expression_values_collapsed_to_symbols.class.cls #double_gel_8d_versus_double_gel_48h.class.cls #double_gel_8d_versus_double_gel_48h_repos |
| Phenotype | class.cls#double_gel_8d_versus_double_gel_48h_repos |
| Upregulated in class | double_gel_48h |
| GeneSet | KCCGNSWTTT_UNKNOWN |
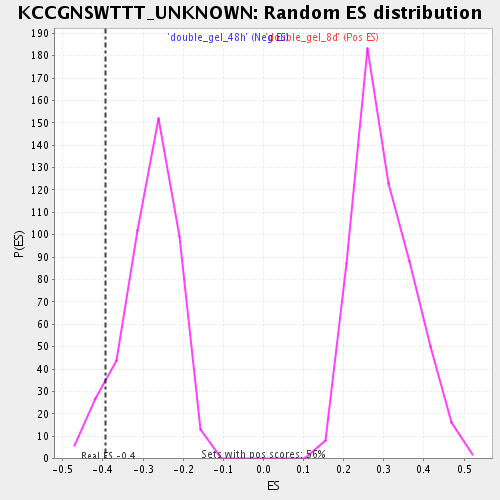
| Enrichment Score (ES) | -0.39269432 |
| Normalized Enrichment Score (NES) | -1.3903333 |
| Nominal p-value | 0.072234765 |
| FDR q-value | 0.34949335 |
| FWER p-Value | 1.0 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | ARMCX2 | ARMCX2 Entrez, Source | armadillo repeat containing, X-linked 2 | 20 | 0.787 | 0.1263 | No |
| 2 | RGL2 | RGL2 Entrez, Source | ral guanine nucleotide dissociation stimulator-like 2 | 439 | 0.247 | 0.1350 | No |
| 3 | SPRED2 | SPRED2 Entrez, Source | sprouty-related, EVH1 domain containing 2 | 528 | 0.221 | 0.1643 | No |
| 4 | POU2F1 | POU2F1 Entrez, Source | POU domain, class 2, transcription factor 1 | 797 | 0.170 | 0.1718 | No |
| 5 | MAX | MAX Entrez, Source | MYC associated factor X | 1040 | 0.142 | 0.1767 | No |
| 6 | PRICKLE1 | PRICKLE1 Entrez, Source | prickle homolog 1 (Drosophila) | 1377 | 0.115 | 0.1701 | No |
| 7 | OTX2 | OTX2 Entrez, Source | orthodenticle homolog 2 (Drosophila) | 2173 | 0.078 | 0.1229 | No |
| 8 | FAAH | FAAH Entrez, Source | fatty acid amide hydrolase | 2484 | 0.069 | 0.1108 | No |
| 9 | ZFP91 | ZFP91 Entrez, Source | zinc finger protein 91 homolog (mouse) | 3065 | 0.053 | 0.0757 | No |
| 10 | MLL5 | MLL5 Entrez, Source | myeloid/lymphoid or mixed-lineage leukemia 5 (trithorax homolog, Drosophila) | 3353 | 0.046 | 0.0616 | No |
| 11 | FOXA2 | FOXA2 Entrez, Source | forkhead box A2 | 3659 | 0.040 | 0.0450 | No |
| 12 | BCL11A | BCL11A Entrez, Source | B-cell CLL/lymphoma 11A (zinc finger protein) | 4147 | 0.030 | 0.0133 | No |
| 13 | UBC | UBC Entrez, Source | ubiquitin C | 4954 | 0.016 | -0.0448 | No |
| 14 | PICALM | PICALM Entrez, Source | phosphatidylinositol binding clathrin assembly protein | 5636 | 0.005 | -0.0952 | No |
| 15 | ENSA | ENSA Entrez, Source | endosulfine alpha | 5749 | 0.003 | -0.1031 | No |
| 16 | TDRD3 | TDRD3 Entrez, Source | tudor domain containing 3 | 6012 | -0.000 | -0.1228 | No |
| 17 | SP1 | SP1 Entrez, Source | Sp1 transcription factor | 6622 | -0.009 | -0.1671 | No |
| 18 | DMTF1 | DMTF1 Entrez, Source | cyclin D binding myb-like transcription factor 1 | 6628 | -0.009 | -0.1659 | No |
| 19 | CBX3 | CBX3 Entrez, Source | chromobox homolog 3 (HP1 gamma homolog, Drosophila) | 6693 | -0.011 | -0.1690 | No |
| 20 | CXXC5 | CXXC5 Entrez, Source | CXXC finger 5 | 6698 | -0.011 | -0.1676 | No |
| 21 | CTCF | CTCF Entrez, Source | CCCTC-binding factor (zinc finger protein) | 6862 | -0.013 | -0.1777 | No |
| 22 | LPHN3 | LPHN3 Entrez, Source | latrophilin 3 | 6892 | -0.013 | -0.1777 | No |
| 23 | CRK | CRK Entrez, Source | v-crk sarcoma virus CT10 oncogene homolog (avian) | 7199 | -0.018 | -0.1978 | No |
| 24 | EIF4A2 | EIF4A2 Entrez, Source | eukaryotic translation initiation factor 4A, isoform 2 | 7219 | -0.018 | -0.1962 | No |
| 25 | CUGBP1 | CUGBP1 Entrez, Source | CUG triplet repeat, RNA binding protein 1 | 7294 | -0.019 | -0.1986 | No |
| 26 | NCAM1 | NCAM1 Entrez, Source | neural cell adhesion molecule 1 | 7426 | -0.021 | -0.2050 | No |
| 27 | SYT11 | SYT11 Entrez, Source | synaptotagmin XI | 7754 | -0.027 | -0.2253 | No |
| 28 | ACLY | ACLY Entrez, Source | ATP citrate lyase | 7774 | -0.027 | -0.2223 | No |
| 29 | DDX5 | DDX5 Entrez, Source | DEAD (Asp-Glu-Ala-Asp) box polypeptide 5 | 8037 | -0.031 | -0.2369 | No |
| 30 | DDX17 | DDX17 Entrez, Source | DEAD (Asp-Glu-Ala-Asp) box polypeptide 17 | 8274 | -0.035 | -0.2489 | No |
| 31 | AP3D1 | AP3D1 Entrez, Source | adaptor-related protein complex 3, delta 1 subunit | 8568 | -0.040 | -0.2645 | No |
| 32 | YWHAE | YWHAE Entrez, Source | tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, epsilon polypeptide | 8841 | -0.044 | -0.2778 | No |
| 33 | CCNA2 | CCNA2 Entrez, Source | cyclin A2 | 8927 | -0.046 | -0.2767 | No |
| 34 | LNPEP | LNPEP Entrez, Source | leucyl/cystinyl aminopeptidase | 8978 | -0.047 | -0.2728 | No |
| 35 | EIF4G2 | EIF4G2 Entrez, Source | eukaryotic translation initiation factor 4 gamma, 2 | 9796 | -0.062 | -0.3242 | No |
| 36 | PPP2R2B | PPP2R2B Entrez, Source | protein phosphatase 2 (formerly 2A), regulatory subunit B (PR 52), beta isoform | 10000 | -0.066 | -0.3287 | No |
| 37 | DAP3 | DAP3 Entrez, Source | death associated protein 3 | 10562 | -0.079 | -0.3581 | No |
| 38 | AKAP8 | AKAP8 Entrez, Source | A kinase (PRKA) anchor protein 8 | 10573 | -0.079 | -0.3459 | No |
| 39 | AP1G1 | AP1G1 Entrez, Source | adaptor-related protein complex 1, gamma 1 subunit | 10665 | -0.082 | -0.3395 | No |
| 40 | HNRPK | HNRPK Entrez, Source | heterogeneous nuclear ribonucleoprotein K | 10815 | -0.086 | -0.3368 | No |
| 41 | PAFAH1B1 | PAFAH1B1 Entrez, Source | platelet-activating factor acetylhydrolase, isoform Ib, alpha subunit 45kDa | 10930 | -0.089 | -0.3310 | No |
| 42 | ATP5G2 | ATP5G2 Entrez, Source | ATP synthase, H+ transporting, mitochondrial F0 complex, subunit C2 (subunit 9) | 11226 | -0.096 | -0.3375 | No |
| 43 | OGT | OGT Entrez, Source | O-linked N-acetylglucosamine (GlcNAc) transferase (UDP-N-acetylglucosamine:polypeptide-N-acetylglucosaminyl transferase) | 11749 | -0.115 | -0.3581 | No |
| 44 | CSNK1A1 | CSNK1A1 Entrez, Source | casein kinase 1, alpha 1 | 12054 | -0.129 | -0.3600 | No |
| 45 | PTMA | PTMA Entrez, Source | prothymosin, alpha (gene sequence 28) | 12489 | -0.154 | -0.3677 | Yes |
| 46 | SFRS10 | SFRS10 Entrez, Source | splicing factor, arginine/serine-rich 10 (transformer 2 homolog, Drosophila) | 12549 | -0.158 | -0.3464 | Yes |
| 47 | PTBP2 | PTBP2 Entrez, Source | polypyrimidine tract binding protein 2 | 12573 | -0.160 | -0.3221 | Yes |
| 48 | USP3 | USP3 Entrez, Source | ubiquitin specific peptidase 3 | 12588 | -0.162 | -0.2969 | Yes |
| 49 | ADNP | ADNP Entrez, Source | activity-dependent neuroprotector | 12610 | -0.164 | -0.2718 | Yes |
| 50 | CRKL | CRKL Entrez, Source | v-crk sarcoma virus CT10 oncogene homolog (avian)-like | 12628 | -0.166 | -0.2462 | Yes |
| 51 | SYNCRIP | SYNCRIP Entrez, Source | synaptotagmin binding, cytoplasmic RNA interacting protein | 12730 | -0.176 | -0.2251 | Yes |
| 52 | HNRPH1 | HNRPH1 Entrez, Source | heterogeneous nuclear ribonucleoprotein H1 (H) | 12830 | -0.189 | -0.2018 | Yes |
| 53 | XPO1 | XPO1 Entrez, Source | exportin 1 (CRM1 homolog, yeast) | 12994 | -0.216 | -0.1789 | Yes |
| 54 | GNG11 | GNG11 Entrez, Source | guanine nucleotide binding protein (G protein), gamma 11 | 13005 | -0.219 | -0.1440 | Yes |
| 55 | ILF3 | ILF3 Entrez, Source | interleukin enhancer binding factor 3, 90kDa | 13043 | -0.231 | -0.1093 | Yes |
| 56 | ASCL1 | ASCL1 Entrez, Source | achaete-scute complex-like 1 (Drosophila) | 13333 | -0.811 | 0.0007 | Yes |