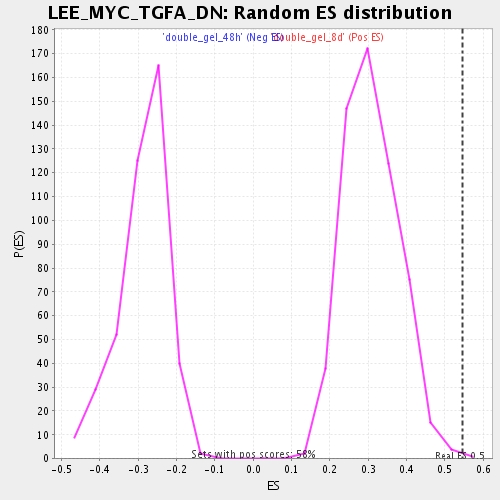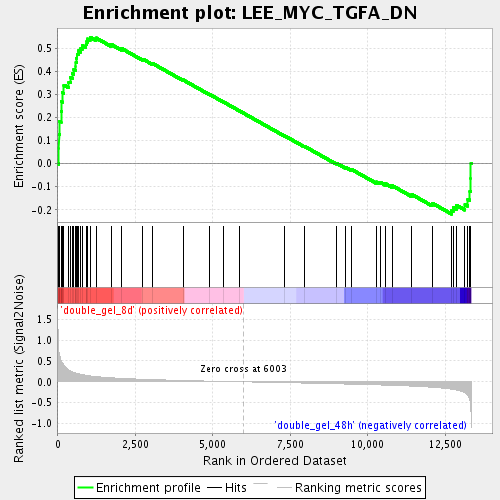
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | double_gel_and_single_gel_rma_expression_values_collapsed_to_symbols.class.cls #double_gel_8d_versus_double_gel_48h.class.cls #double_gel_8d_versus_double_gel_48h_repos |
| Phenotype | class.cls#double_gel_8d_versus_double_gel_48h_repos |
| Upregulated in class | double_gel_8d |
| GeneSet | LEE_MYC_TGFA_DN |
| Enrichment Score (ES) | 0.54650134 |
| Normalized Enrichment Score (NES) | 1.7720478 |
| Nominal p-value | 0.0017301039 |
| FDR q-value | 0.030658634 |
| FWER p-Value | 0.987 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | HLA-DMA | HLA-DMA Entrez, Source | major histocompatibility complex, class II, DM alpha | 32 | 0.725 | 0.0633 | Yes |
| 2 | PCK1 | PCK1 Entrez, Source | phosphoenolpyruvate carboxykinase 1 (soluble) | 35 | 0.711 | 0.1277 | Yes |
| 3 | G6PC | G6PC Entrez, Source | glucose-6-phosphatase, catalytic subunit | 57 | 0.628 | 0.1831 | Yes |
| 4 | RGS16 | RGS16 Entrez, Source | regulator of G-protein signalling 16 | 103 | 0.505 | 0.2255 | Yes |
| 5 | FABP2 | FABP2 Entrez, Source | fatty acid binding protein 2, intestinal | 121 | 0.476 | 0.2674 | Yes |
| 6 | GOT1 | GOT1 Entrez, Source | glutamic-oxaloacetic transaminase 1, soluble (aspartate aminotransferase 1) | 144 | 0.453 | 0.3068 | Yes |
| 7 | LIPC | LIPC Entrez, Source | lipase, hepatic | 193 | 0.397 | 0.3392 | Yes |
| 8 | CPT2 | CPT2 Entrez, Source | carnitine palmitoyltransferase II | 351 | 0.281 | 0.3528 | Yes |
| 9 | PXMP2 | PXMP2 Entrez, Source | peroxisomal membrane protein 2, 22kDa | 408 | 0.257 | 0.3719 | Yes |
| 10 | KLF15 | KLF15 Entrez, Source | Kruppel-like factor 15 | 466 | 0.240 | 0.3894 | Yes |
| 11 | TDO2 | TDO2 Entrez, Source | tryptophan 2,3-dioxygenase | 514 | 0.223 | 0.4062 | Yes |
| 12 | FABP1 | FABP1 Entrez, Source | fatty acid binding protein 1, liver | 580 | 0.210 | 0.4203 | Yes |
| 13 | CA5A | CA5A Entrez, Source | carbonic anhydrase VA, mitochondrial | 582 | 0.210 | 0.4393 | Yes |
| 14 | SLC25A22 | SLC25A22 Entrez, Source | solute carrier family 25 (mitochondrial carrier: glutamate), member 22 | 605 | 0.206 | 0.4563 | Yes |
| 15 | IL1RAP | IL1RAP Entrez, Source | interleukin 1 receptor accessory protein | 616 | 0.203 | 0.4739 | Yes |
| 16 | BHMT | BHMT Entrez, Source | betaine-homocysteine methyltransferase | 669 | 0.192 | 0.4874 | Yes |
| 17 | ALDH1A1 | ALDH1A1 Entrez, Source | aldehyde dehydrogenase 1 family, member A1 | 739 | 0.178 | 0.4984 | Yes |
| 18 | HADH2 | HADH2 Entrez, Source | hydroxyacyl-Coenzyme A dehydrogenase, type II | 782 | 0.173 | 0.5109 | Yes |
| 19 | BAAT | BAAT Entrez, Source | bile acid Coenzyme A: amino acid N-acyltransferase (glycine N-choloyltransferase) | 906 | 0.156 | 0.5157 | Yes |
| 20 | KYNU | KYNU Entrez, Source | kynureninase (L-kynurenine hydrolase) | 934 | 0.153 | 0.5276 | Yes |
| 21 | EPHX2 | EPHX2 Entrez, Source | epoxide hydrolase 2, cytoplasmic | 964 | 0.151 | 0.5391 | Yes |
| 22 | OTC | OTC Entrez, Source | ornithine carbamoyltransferase | 1038 | 0.143 | 0.5465 | Yes |
| 23 | MAPKAPK2 | MAPKAPK2 Entrez, Source | mitogen-activated protein kinase-activated protein kinase 2 | 1230 | 0.125 | 0.5435 | No |
| 24 | HSD11B1 | HSD11B1 Entrez, Source | hydroxysteroid (11-beta) dehydrogenase 1 | 1720 | 0.096 | 0.5155 | No |
| 25 | ALAS2 | ALAS2 Entrez, Source | aminolevulinate, delta-, synthase 2 (sideroblastic/hypochromic anemia) | 2058 | 0.083 | 0.4976 | No |
| 26 | ACOX1 | ACOX1 Entrez, Source | acyl-Coenzyme A oxidase 1, palmitoyl | 2738 | 0.062 | 0.4521 | No |
| 27 | TRPV2 | TRPV2 Entrez, Source | transient receptor potential cation channel, subfamily V, member 2 | 3055 | 0.053 | 0.4332 | No |
| 28 | ELOVL5 | ELOVL5 Entrez, Source | ELOVL family member 5, elongation of long chain fatty acids (FEN1/Elo2, SUR4/Elo3-like, yeast) | 4037 | 0.032 | 0.3623 | No |
| 29 | HPD | HPD Entrez, Source | 4-hydroxyphenylpyruvate dioxygenase | 4885 | 0.017 | 0.3001 | No |
| 30 | OAT | OAT Entrez, Source | ornithine aminotransferase (gyrate atrophy) | 5350 | 0.009 | 0.2660 | No |
| 31 | CYP7B1 | CYP7B1 Entrez, Source | cytochrome P450, family 7, subfamily B, polypeptide 1 | 5862 | 0.002 | 0.2278 | No |
| 32 | ABAT | ABAT Entrez, Source | 4-aminobutyrate aminotransferase | 7326 | -0.020 | 0.1195 | No |
| 33 | IGF1 | IGF1 Entrez, Source | insulin-like growth factor 1 (somatomedin C) | 7950 | -0.030 | 0.0754 | No |
| 34 | CMAH | CMAH Entrez, Source | cytidine monophosphate-N-acetylneuraminic acid hydroxylase (CMP-N-acetylneuraminate monooxygenase) | 8981 | -0.047 | 0.0022 | No |
| 35 | PSEN2 | PSEN2 Entrez, Source | presenilin 2 (Alzheimer disease 4) | 9275 | -0.052 | -0.0151 | No |
| 36 | NR1H3 | NR1H3 Entrez, Source | nuclear receptor subfamily 1, group H, member 3 | 9459 | -0.055 | -0.0239 | No |
| 37 | CRISP2 | CRISP2 Entrez, Source | cysteine-rich secretory protein 2 | 10285 | -0.072 | -0.0794 | No |
| 38 | GPR37 | GPR37 Entrez, Source | G protein-coupled receptor 37 (endothelin receptor type B-like) | 10399 | -0.075 | -0.0811 | No |
| 39 | CYP4F2 | CYP4F2 Entrez, Source | cytochrome P450, family 4, subfamily F, polypeptide 2 | 10574 | -0.079 | -0.0870 | No |
| 40 | C4BPA | C4BPA Entrez, Source | complement component 4 binding protein, alpha | 10785 | -0.085 | -0.0951 | No |
| 41 | AQP8 | AQP8 Entrez, Source | aquaporin 8 | 11422 | -0.103 | -0.1336 | No |
| 42 | HAL | HAL Entrez, Source | histidine ammonia-lyase | 12080 | -0.131 | -0.1712 | No |
| 43 | ABCG2 | ABCG2 Entrez, Source | ATP-binding cassette, sub-family G (WHITE), member 2 | 12709 | -0.174 | -0.2026 | No |
| 44 | KHK | KHK Entrez, Source | ketohexokinase (fructokinase) | 12757 | -0.178 | -0.1900 | No |
| 45 | EGFR | EGFR Entrez, Source | epidermal growth factor receptor (erythroblastic leukemia viral (v-erb-b) oncogene homolog, avian) | 12857 | -0.193 | -0.1800 | No |
| 46 | G0S2 | G0S2 Entrez, Source | G0/G1switch 2 | 13137 | -0.265 | -0.1769 | No |
| 47 | IGFALS | IGFALS Entrez, Source | insulin-like growth factor binding protein, acid labile subunit | 13230 | -0.325 | -0.1544 | No |
| 48 | SLC27A5 | SLC27A5 Entrez, Source | solute carrier family 27 (fatty acid transporter), member 5 | 13297 | -0.441 | -0.1194 | No |
| 49 | CA3 | CA3 Entrez, Source | carbonic anhydrase III, muscle specific | 13327 | -0.634 | -0.0641 | No |
| 50 | RGN | RGN Entrez, Source | regucalcin (senescence marker protein-30) | 13331 | -0.718 | 0.0008 | No |