Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | double_gel_and_single_gel_rma_expression_values_collapsed_to_symbols.class.cls #double_gel_8d_versus_double_gel_48h.class.cls #double_gel_8d_versus_double_gel_48h_repos |
| Phenotype | class.cls#double_gel_8d_versus_double_gel_48h_repos |
| Upregulated in class | double_gel_48h |
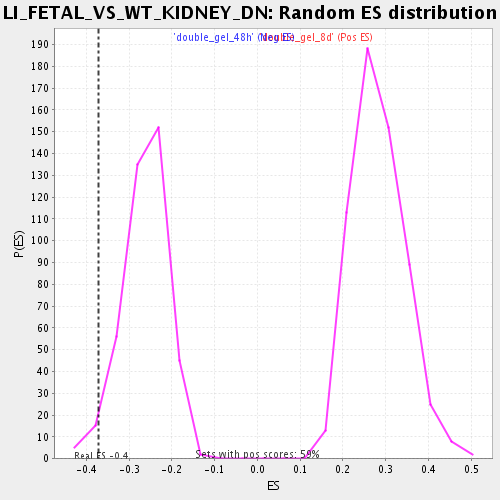
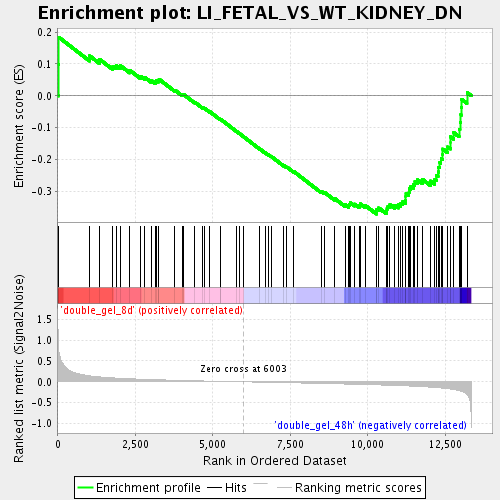
| GeneSet | LI_FETAL_VS_WT_KIDNEY_DN |
| Enrichment Score (ES) | -0.37217593 |
| Normalized Enrichment Score (NES) | -1.4153543 |
| Nominal p-value | 0.026829269 |
| FDR q-value | 0.33463013 |
| FWER p-Value | 1.0 |

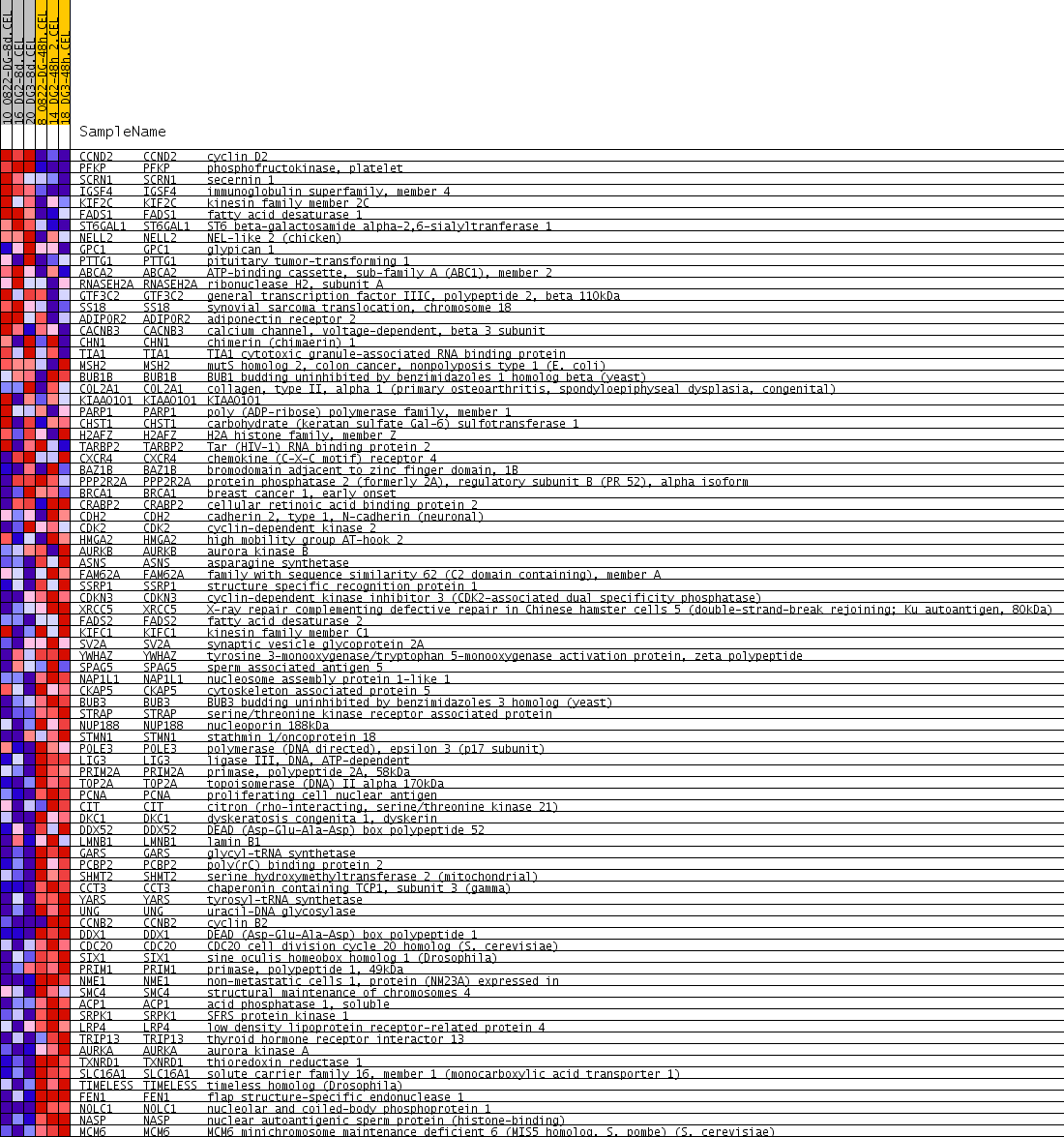
| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | CCND2 | CCND2 Entrez, Source | cyclin D2 | 10 | 0.885 | 0.0999 | No |
| 2 | PFKP | PFKP Entrez, Source | phosphofructokinase, platelet | 29 | 0.753 | 0.1841 | No |
| 3 | SCRN1 | SCRN1 Entrez, Source | secernin 1 | 1023 | 0.145 | 0.1257 | No |
| 4 | IGSF4 | IGSF4 Entrez, Source | immunoglobulin superfamily, member 4 | 1345 | 0.117 | 0.1147 | No |
| 5 | KIF2C | KIF2C Entrez, Source | kinesin family member 2C | 1773 | 0.094 | 0.0932 | No |
| 6 | FADS1 | FADS1 Entrez, Source | fatty acid desaturase 1 | 1887 | 0.089 | 0.0948 | No |
| 7 | ST6GAL1 | ST6GAL1 Entrez, Source | ST6 beta-galactosamide alpha-2,6-sialyltranferase 1 | 2013 | 0.085 | 0.0950 | No |
| 8 | NELL2 | NELL2 Entrez, Source | NEL-like 2 (chicken) | 2324 | 0.073 | 0.0799 | No |
| 9 | GPC1 | GPC1 Entrez, Source | glypican 1 | 2672 | 0.063 | 0.0609 | No |
| 10 | PTTG1 | PTTG1 Entrez, Source | pituitary tumor-transforming 1 | 2806 | 0.060 | 0.0577 | No |
| 11 | ABCA2 | ABCA2 Entrez, Source | ATP-binding cassette, sub-family A (ABC1), member 2 | 3022 | 0.054 | 0.0476 | No |
| 12 | RNASEH2A | RNASEH2A Entrez, Source | ribonuclease H2, subunit A | 3157 | 0.051 | 0.0432 | No |
| 13 | GTF3C2 | GTF3C2 Entrez, Source | general transcription factor IIIC, polypeptide 2, beta 110kDa | 3171 | 0.050 | 0.0480 | No |
| 14 | SS18 | SS18 Entrez, Source | synovial sarcoma translocation, chromosome 18 | 3236 | 0.049 | 0.0487 | No |
| 15 | ADIPOR2 | ADIPOR2 Entrez, Source | adiponectin receptor 2 | 3261 | 0.048 | 0.0523 | No |
| 16 | CACNB3 | CACNB3 Entrez, Source | calcium channel, voltage-dependent, beta 3 subunit | 3774 | 0.037 | 0.0179 | No |
| 17 | CHN1 | CHN1 Entrez, Source | chimerin (chimaerin) 1 | 4021 | 0.032 | 0.0030 | No |
| 18 | TIA1 | TIA1 Entrez, Source | TIA1 cytotoxic granule-associated RNA binding protein | 4046 | 0.032 | 0.0049 | No |
| 19 | MSH2 | MSH2 Entrez, Source | mutS homolog 2, colon cancer, nonpolyposis type 1 (E. coli) | 4414 | 0.025 | -0.0200 | No |
| 20 | BUB1B | BUB1B Entrez, Source | BUB1 budding uninhibited by benzimidazoles 1 homolog beta (yeast) | 4680 | 0.020 | -0.0376 | No |
| 21 | COL2A1 | COL2A1 Entrez, Source | collagen, type II, alpha 1 (primary osteoarthritis, spondyloepiphyseal dysplasia, congenital) | 4726 | 0.020 | -0.0388 | No |
| 22 | KIAA0101 | KIAA0101 Entrez, Source | KIAA0101 | 4903 | 0.017 | -0.0502 | No |
| 23 | PARP1 | PARP1 Entrez, Source | poly (ADP-ribose) polymerase family, member 1 | 5231 | 0.011 | -0.0735 | No |
| 24 | CHST1 | CHST1 Entrez, Source | carbohydrate (keratan sulfate Gal-6) sulfotransferase 1 | 5261 | 0.011 | -0.0745 | No |
| 25 | H2AFZ | H2AFZ Entrez, Source | H2A histone family, member Z | 5766 | 0.003 | -0.1122 | No |
| 26 | TARBP2 | TARBP2 Entrez, Source | Tar (HIV-1) RNA binding protein 2 | 5848 | 0.002 | -0.1180 | No |
| 27 | CXCR4 | CXCR4 Entrez, Source | chemokine (C-X-C motif) receptor 4 | 5981 | 0.000 | -0.1279 | No |
| 28 | BAZ1B | BAZ1B Entrez, Source | bromodomain adjacent to zinc finger domain, 1B | 6516 | -0.008 | -0.1673 | No |
| 29 | PPP2R2A | PPP2R2A Entrez, Source | protein phosphatase 2 (formerly 2A), regulatory subunit B (PR 52), alpha isoform | 6690 | -0.011 | -0.1792 | No |
| 30 | BRCA1 | BRCA1 Entrez, Source | breast cancer 1, early onset | 6790 | -0.012 | -0.1853 | No |
| 31 | CRABP2 | CRABP2 Entrez, Source | cellular retinoic acid binding protein 2 | 6879 | -0.013 | -0.1904 | No |
| 32 | CDH2 | CDH2 Entrez, Source | cadherin 2, type 1, N-cadherin (neuronal) | 7290 | -0.019 | -0.2191 | No |
| 33 | CDK2 | CDK2 Entrez, Source | cyclin-dependent kinase 2 | 7386 | -0.021 | -0.2239 | No |
| 34 | HMGA2 | HMGA2 Entrez, Source | high mobility group AT-hook 2 | 7610 | -0.024 | -0.2380 | No |
| 35 | AURKB | AURKB Entrez, Source | aurora kinase B | 8506 | -0.039 | -0.3011 | No |
| 36 | ASNS | ASNS Entrez, Source | asparagine synthetase | 8596 | -0.040 | -0.3032 | No |
| 37 | FAM62A | FAM62A Entrez, Source | family with sequence similarity 62 (C2 domain containing), member A | 8936 | -0.046 | -0.3235 | No |
| 38 | SSRP1 | SSRP1 Entrez, Source | structure specific recognition protein 1 | 9266 | -0.052 | -0.3424 | No |
| 39 | CDKN3 | CDKN3 Entrez, Source | cyclin-dependent kinase inhibitor 3 (CDK2-associated dual specificity phosphatase) | 9384 | -0.054 | -0.3451 | No |
| 40 | XRCC5 | XRCC5 Entrez, Source | X-ray repair complementing defective repair in Chinese hamster cells 5 (double-strand-break rejoining; Ku autoantigen, 80kDa) | 9405 | -0.054 | -0.3405 | No |
| 41 | FADS2 | FADS2 Entrez, Source | fatty acid desaturase 2 | 9436 | -0.055 | -0.3365 | No |
| 42 | KIFC1 | KIFC1 Entrez, Source | kinesin family member C1 | 9575 | -0.057 | -0.3404 | No |
| 43 | SV2A | SV2A Entrez, Source | synaptic vesicle glycoprotein 2A | 9725 | -0.061 | -0.3447 | No |
| 44 | YWHAZ | YWHAZ Entrez, Source | tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, zeta polypeptide | 9753 | -0.061 | -0.3398 | No |
| 45 | SPAG5 | SPAG5 Entrez, Source | sperm associated antigen 5 | 9912 | -0.065 | -0.3444 | No |
| 46 | NAP1L1 | NAP1L1 Entrez, Source | nucleosome assembly protein 1-like 1 | 10281 | -0.072 | -0.3640 | Yes |
| 47 | CKAP5 | CKAP5 Entrez, Source | cytoskeleton associated protein 5 | 10289 | -0.072 | -0.3563 | Yes |
| 48 | BUB3 | BUB3 Entrez, Source | BUB3 budding uninhibited by benzimidazoles 3 homolog (yeast) | 10355 | -0.074 | -0.3528 | Yes |
| 49 | STRAP | STRAP Entrez, Source | serine/threonine kinase receptor associated protein | 10601 | -0.080 | -0.3622 | Yes |
| 50 | NUP188 | NUP188 Entrez, Source | nucleoporin 188kDa | 10620 | -0.080 | -0.3545 | Yes |
| 51 | STMN1 | STMN1 Entrez, Source | stathmin 1/oncoprotein 18 | 10649 | -0.081 | -0.3474 | Yes |
| 52 | POLE3 | POLE3 Entrez, Source | polymerase (DNA directed), epsilon 3 (p17 subunit) | 10715 | -0.083 | -0.3428 | Yes |
| 53 | LIG3 | LIG3 Entrez, Source | ligase III, DNA, ATP-dependent | 10864 | -0.087 | -0.3441 | Yes |
| 54 | PRIM2A | PRIM2A Entrez, Source | primase, polypeptide 2A, 58kDa | 10985 | -0.090 | -0.3429 | Yes |
| 55 | TOP2A | TOP2A Entrez, Source | topoisomerase (DNA) II alpha 170kDa | 11066 | -0.092 | -0.3385 | Yes |
| 56 | PCNA | PCNA Entrez, Source | proliferating cell nuclear antigen | 11131 | -0.094 | -0.3326 | Yes |
| 57 | CIT | CIT Entrez, Source | citron (rho-interacting, serine/threonine kinase 21) | 11218 | -0.096 | -0.3281 | Yes |
| 58 | DKC1 | DKC1 Entrez, Source | dyskeratosis congenita 1, dyskerin | 11229 | -0.096 | -0.3179 | Yes |
| 59 | DDX52 | DDX52 Entrez, Source | DEAD (Asp-Glu-Ala-Asp) box polypeptide 52 | 11233 | -0.097 | -0.3072 | Yes |
| 60 | LMNB1 | LMNB1 Entrez, Source | lamin B1 | 11330 | -0.100 | -0.3031 | Yes |
| 61 | GARS | GARS Entrez, Source | glycyl-tRNA synthetase | 11341 | -0.100 | -0.2925 | Yes |
| 62 | PCBP2 | PCBP2 Entrez, Source | poly(rC) binding protein 2 | 11388 | -0.102 | -0.2844 | Yes |
| 63 | SHMT2 | SHMT2 Entrez, Source | serine hydroxymethyltransferase 2 (mitochondrial) | 11474 | -0.105 | -0.2789 | Yes |
| 64 | CCT3 | CCT3 Entrez, Source | chaperonin containing TCP1, subunit 3 (gamma) | 11522 | -0.106 | -0.2703 | Yes |
| 65 | YARS | YARS Entrez, Source | tyrosyl-tRNA synthetase | 11607 | -0.110 | -0.2642 | Yes |
| 66 | UNG | UNG Entrez, Source | uracil-DNA glycosylase | 11765 | -0.116 | -0.2629 | Yes |
| 67 | CCNB2 | CCNB2 Entrez, Source | cyclin B2 | 12025 | -0.128 | -0.2679 | Yes |
| 68 | DDX1 | DDX1 Entrez, Source | DEAD (Asp-Glu-Ala-Asp) box polypeptide 1 | 12162 | -0.135 | -0.2629 | Yes |
| 69 | CDC20 | CDC20 Entrez, Source | CDC20 cell division cycle 20 homolog (S. cerevisiae) | 12218 | -0.137 | -0.2514 | Yes |
| 70 | SIX1 | SIX1 Entrez, Source | sine oculis homeobox homolog 1 (Drosophila) | 12271 | -0.141 | -0.2393 | Yes |
| 71 | PRIM1 | PRIM1 Entrez, Source | primase, polypeptide 1, 49kDa | 12284 | -0.141 | -0.2242 | Yes |
| 72 | NME1 | NME1 Entrez, Source | non-metastatic cells 1, protein (NM23A) expressed in | 12324 | -0.144 | -0.2108 | Yes |
| 73 | SMC4 | SMC4 Entrez, Source | structural maintenance of chromosomes 4 | 12364 | -0.147 | -0.1970 | Yes |
| 74 | ACP1 | ACP1 Entrez, Source | acid phosphatase 1, soluble | 12417 | -0.149 | -0.1840 | Yes |
| 75 | SRPK1 | SRPK1 Entrez, Source | SFRS protein kinase 1 | 12424 | -0.150 | -0.1674 | Yes |
| 76 | LRP4 | LRP4 Entrez, Source | low density lipoprotein receptor-related protein 4 | 12582 | -0.161 | -0.1610 | Yes |
| 77 | TRIP13 | TRIP13 Entrez, Source | thyroid hormone receptor interactor 13 | 12670 | -0.170 | -0.1482 | Yes |
| 78 | AURKA | AURKA Entrez, Source | aurora kinase A | 12673 | -0.170 | -0.1290 | Yes |
| 79 | TXNRD1 | TXNRD1 Entrez, Source | thioredoxin reductase 1 | 12775 | -0.181 | -0.1160 | Yes |
| 80 | SLC16A1 | SLC16A1 Entrez, Source | solute carrier family 16, member 1 (monocarboxylic acid transporter 1) | 12950 | -0.209 | -0.1054 | Yes |
| 81 | TIMELESS | TIMELESS Entrez, Source | timeless homolog (Drosophila) | 12978 | -0.214 | -0.0831 | Yes |
| 82 | FEN1 | FEN1 Entrez, Source | flap structure-specific endonuclease 1 | 13004 | -0.219 | -0.0601 | Yes |
| 83 | NOLC1 | NOLC1 Entrez, Source | nucleolar and coiled-body phosphoprotein 1 | 13015 | -0.221 | -0.0357 | Yes |
| 84 | NASP | NASP Entrez, Source | nuclear autoantigenic sperm protein (histone-binding) | 13031 | -0.226 | -0.0111 | Yes |
| 85 | MCM6 | MCM6 Entrez, Source | MCM6 minichromosome maintenance deficient 6 (MIS5 homolog, S. pombe) (S. cerevisiae) | 13204 | -0.304 | 0.0104 | Yes |