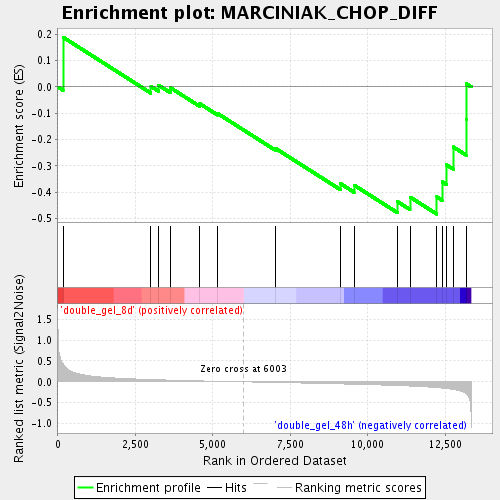
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | double_gel_and_single_gel_rma_expression_values_collapsed_to_symbols.class.cls #double_gel_8d_versus_double_gel_48h.class.cls #double_gel_8d_versus_double_gel_48h_repos |
| Phenotype | class.cls#double_gel_8d_versus_double_gel_48h_repos |
| Upregulated in class | double_gel_48h |
| GeneSet | MARCINIAK_CHOP_DIFF |
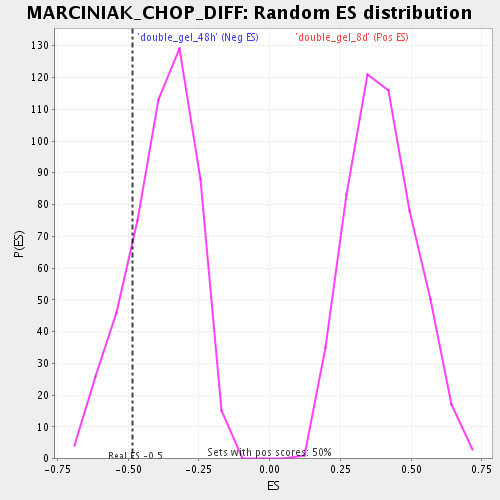
| Enrichment Score (ES) | -0.483572 |
| Normalized Enrichment Score (NES) | -1.2754947 |
| Nominal p-value | 0.17943548 |
| FDR q-value | 0.5143352 |
| FWER p-Value | 1.0 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | AMPD3 | AMPD3 Entrez, Source | adenosine monophosphate deaminase (isoform E) | 167 | 0.426 | 0.1892 | No |
| 2 | WFS1 | WFS1 Entrez, Source | Wolfram syndrome 1 (wolframin) | 3003 | 0.054 | 0.0022 | No |
| 3 | ADH7 | ADH7 Entrez, Source | alcohol dehydrogenase 7 (class IV), mu or sigma polypeptide | 3254 | 0.048 | 0.0062 | No |
| 4 | MYD116 | 3627 | 0.040 | -0.0026 | No | ||
| 5 | PARD6A | PARD6A Entrez, Source | par-6 partitioning defective 6 homolog alpha (C.elegans) | 4580 | 0.022 | -0.0637 | No |
| 6 | CHKA | CHKA Entrez, Source | choline kinase alpha | 5155 | 0.012 | -0.1009 | No |
| 7 | KITLG | KITLG Entrez, Source | KIT ligand | 7033 | -0.016 | -0.2344 | No |
| 8 | PPAN | PPAN Entrez, Source | peter pan homolog (Drosophila) | 9124 | -0.049 | -0.3679 | No |
| 9 | ERO1L | ERO1L Entrez, Source | ERO1-like (S. cerevisiae) | 9559 | -0.057 | -0.3735 | No |
| 10 | POLR1A | POLR1A Entrez, Source | polymerase (RNA) I polypeptide A, 194kDa | 10957 | -0.089 | -0.4360 | Yes |
| 11 | TCEA1 | TCEA1 Entrez, Source | transcription elongation factor A (SII), 1 | 11364 | -0.101 | -0.4187 | Yes |
| 12 | CAR6 | 12229 | -0.138 | -0.4181 | Yes | ||
| 13 | CLCN3 | CLCN3 Entrez, Source | chloride channel 3 | 12400 | -0.149 | -0.3606 | Yes |
| 14 | MTM1 | MTM1 Entrez, Source | myotubularin 1 | 12542 | -0.158 | -0.2963 | Yes |
| 15 | CDCA7 | CDCA7 Entrez, Source | cell division cycle associated 7 | 12759 | -0.178 | -0.2280 | Yes |
| 16 | PRSS15 | PRSS15 Entrez, Source | protease, serine, 15 | 13176 | -0.285 | -0.1244 | Yes |
| 17 | DDIT3 | DDIT3 Entrez, Source | DNA-damage-inducible transcript 3 | 13182 | -0.289 | 0.0120 | Yes |