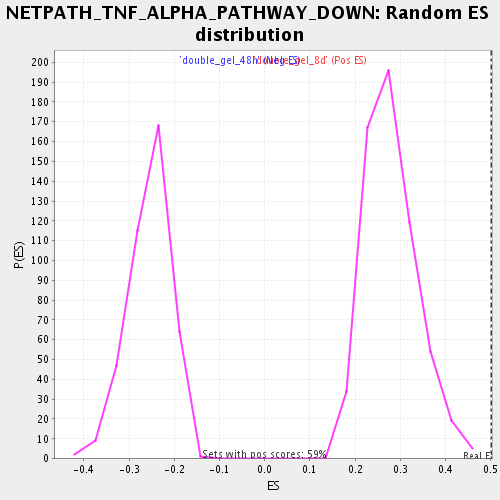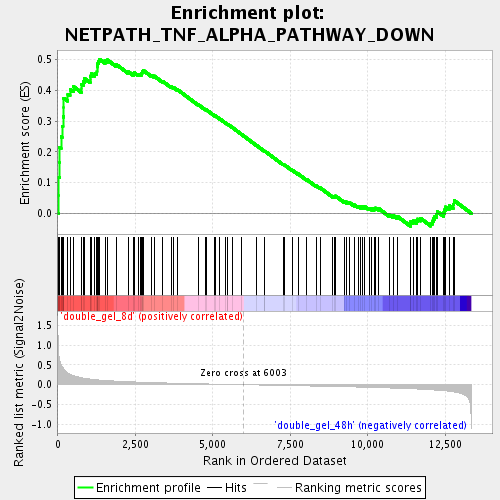
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | double_gel_and_single_gel_rma_expression_values_collapsed_to_symbols.class.cls #double_gel_8d_versus_double_gel_48h.class.cls #double_gel_8d_versus_double_gel_48h_repos |
| Phenotype | class.cls#double_gel_8d_versus_double_gel_48h_repos |
| Upregulated in class | double_gel_8d |
| GeneSet | NETPATH_TNF_ALPHA_PATHWAY_DOWN |
| Enrichment Score (ES) | 0.5009195 |
| Normalized Enrichment Score (NES) | 1.7982394 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.02592502 |
| FWER p-Value | 0.973 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | LDHB | LDHB Entrez, Source | lactate dehydrogenase B | 22 | 0.785 | 0.0592 | Yes |
| 2 | CTGF | CTGF Entrez, Source | connective tissue growth factor | 27 | 0.757 | 0.1176 | Yes |
| 3 | CCL20 | CCL20 Entrez, Source | chemokine (C-C motif) ligand 20 | 50 | 0.641 | 0.1657 | Yes |
| 4 | G6PC | G6PC Entrez, Source | glucose-6-phosphatase, catalytic subunit | 57 | 0.628 | 0.2139 | Yes |
| 5 | RGS16 | RGS16 Entrez, Source | regulator of G-protein signalling 16 | 103 | 0.505 | 0.2497 | Yes |
| 6 | CXCL2 | CXCL2 Entrez, Source | chemokine (C-X-C motif) ligand 2 | 138 | 0.458 | 0.2827 | Yes |
| 7 | ITGB4 | ITGB4 Entrez, Source | integrin, beta 4 | 165 | 0.429 | 0.3140 | Yes |
| 8 | SERPINH1 | SERPINH1 Entrez, Source | serpin peptidase inhibitor, clade H (heat shock protein 47), member 1, (collagen binding protein 1) | 184 | 0.403 | 0.3439 | Yes |
| 9 | ABCG1 | ABCG1 Entrez, Source | ATP-binding cassette, sub-family G (WHITE), member 1 | 187 | 0.400 | 0.3748 | Yes |
| 10 | TYRO3 | TYRO3 Entrez, Source | TYRO3 protein tyrosine kinase | 321 | 0.292 | 0.3874 | Yes |
| 11 | IER3 | IER3 Entrez, Source | immediate early response 3 | 392 | 0.264 | 0.4026 | Yes |
| 12 | DDAH2 | DDAH2 Entrez, Source | dimethylarginine dimethylaminohydrolase 2 | 501 | 0.227 | 0.4120 | Yes |
| 13 | NFKBIA | NFKBIA Entrez, Source | nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor, alpha | 754 | 0.176 | 0.4067 | Yes |
| 14 | EGR1 | EGR1 Entrez, Source | early growth response 1 | 756 | 0.176 | 0.4203 | Yes |
| 15 | CKB | CKB Entrez, Source | creatine kinase, brain | 820 | 0.166 | 0.4284 | Yes |
| 16 | JUNB | JUNB Entrez, Source | jun B proto-oncogene | 861 | 0.161 | 0.4379 | Yes |
| 17 | CXCL1 | CXCL1 Entrez, Source | chemokine (C-X-C motif) ligand 1 (melanoma growth stimulating activity, alpha) | 1041 | 0.142 | 0.4354 | Yes |
| 18 | NR4A1 | NR4A1 Entrez, Source | nuclear receptor subfamily 4, group A, member 1 | 1043 | 0.142 | 0.4463 | Yes |
| 19 | ACSL3 | ACSL3 Entrez, Source | acyl-CoA synthetase long-chain family member 3 | 1086 | 0.137 | 0.4538 | Yes |
| 20 | INSIG1 | INSIG1 Entrez, Source | insulin induced gene 1 | 1195 | 0.128 | 0.4556 | Yes |
| 21 | ZFP36 | ZFP36 Entrez, Source | zinc finger protein 36, C3H type, homolog (mouse) | 1260 | 0.123 | 0.4603 | Yes |
| 22 | CLK1 | CLK1 Entrez, Source | CDC-like kinase 1 | 1262 | 0.123 | 0.4697 | Yes |
| 23 | CLIC1 | CLIC1 Entrez, Source | chloride intracellular channel 1 | 1263 | 0.123 | 0.4793 | Yes |
| 24 | KLF6 | KLF6 Entrez, Source | Kruppel-like factor 6 | 1286 | 0.121 | 0.4870 | Yes |
| 25 | MYH10 | MYH10 Entrez, Source | myosin, heavy chain 10, non-muscle | 1305 | 0.119 | 0.4949 | Yes |
| 26 | AKAP9 | AKAP9 Entrez, Source | A kinase (PRKA) anchor protein (yotiao) 9 | 1346 | 0.117 | 0.5009 | Yes |
| 27 | PLK2 | PLK2 Entrez, Source | polo-like kinase 2 (Drosophila) | 1523 | 0.107 | 0.4959 | No |
| 28 | HADHA | HADHA Entrez, Source | hydroxyacyl-Coenzyme A dehydrogenase/3-ketoacyl-Coenzyme A thiolase/enoyl-Coenzyme A hydratase (trifunctional protein), alpha subunit | 1594 | 0.103 | 0.4986 | No |
| 29 | APOA1 | APOA1 Entrez, Source | apolipoprotein A-I | 1886 | 0.089 | 0.4836 | No |
| 30 | TRADD | TRADD Entrez, Source | TNFRSF1A-associated via death domain | 2264 | 0.075 | 0.4609 | No |
| 31 | ADD3 | ADD3 Entrez, Source | adducin 3 (gamma) | 2434 | 0.070 | 0.4536 | No |
| 32 | VIL2 | VIL2 Entrez, Source | villin 2 (ezrin) | 2459 | 0.069 | 0.4571 | No |
| 33 | FZD2 | FZD2 Entrez, Source | frizzled homolog 2 (Drosophila) | 2586 | 0.065 | 0.4527 | No |
| 34 | CCND1 | CCND1 Entrez, Source | cyclin D1 | 2668 | 0.063 | 0.4514 | No |
| 35 | SIPA1L1 | SIPA1L1 Entrez, Source | signal-induced proliferation-associated 1 like 1 | 2701 | 0.063 | 0.4539 | No |
| 36 | FN1 | FN1 Entrez, Source | fibronectin 1 | 2715 | 0.062 | 0.4577 | No |
| 37 | DUSP2 | DUSP2 Entrez, Source | dual specificity phosphatase 2 | 2724 | 0.062 | 0.4619 | No |
| 38 | EGR3 | EGR3 Entrez, Source | early growth response 3 | 2754 | 0.061 | 0.4645 | No |
| 39 | C1QB | C1QB Entrez, Source | complement component 1, q subcomponent, B chain | 3017 | 0.054 | 0.4489 | No |
| 40 | JUN | JUN Entrez, Source | jun oncogene | 3104 | 0.052 | 0.4464 | No |
| 41 | SDC4 | SDC4 Entrez, Source | syndecan 4 (amphiglycan, ryudocan) | 3385 | 0.045 | 0.4288 | No |
| 42 | ANK1 | ANK1 Entrez, Source | ankyrin 1, erythrocytic | 3656 | 0.040 | 0.4115 | No |
| 43 | NFKB1 | NFKB1 Entrez, Source | nuclear factor of kappa light polypeptide gene enhancer in B-cells 1 (p105) | 3740 | 0.038 | 0.4081 | No |
| 44 | IL6 | IL6 Entrez, Source | interleukin 6 (interferon, beta 2) | 3854 | 0.036 | 0.4024 | No |
| 45 | WASF1 | WASF1 Entrez, Source | WAS protein family, member 1 | 4524 | 0.023 | 0.3536 | No |
| 46 | FASTK | FASTK Entrez, Source | Fas-activated serine/threonine kinase | 4763 | 0.019 | 0.3371 | No |
| 47 | COPB2 | COPB2 Entrez, Source | coatomer protein complex, subunit beta 2 (beta prime) | 4784 | 0.019 | 0.3370 | No |
| 48 | SRPR | SRPR Entrez, Source | signal recognition particle receptor ('docking protein') | 5065 | 0.014 | 0.3170 | No |
| 49 | MARK3 | MARK3 Entrez, Source | MAP/microtubule affinity-regulating kinase 3 | 5075 | 0.014 | 0.3174 | No |
| 50 | NR3C1 | NR3C1 Entrez, Source | nuclear receptor subfamily 3, group C, member 1 (glucocorticoid receptor) | 5227 | 0.011 | 0.3069 | No |
| 51 | SF3B1 | SF3B1 Entrez, Source | splicing factor 3b, subunit 1, 155kDa | 5416 | 0.009 | 0.2933 | No |
| 52 | CD80 | CD80 Entrez, Source | CD80 molecule | 5469 | 0.008 | 0.2900 | No |
| 53 | TFF3 | TFF3 Entrez, Source | trefoil factor 3 (intestinal) | 5480 | 0.008 | 0.2898 | No |
| 54 | PICALM | PICALM Entrez, Source | phosphatidylinositol binding clathrin assembly protein | 5636 | 0.005 | 0.2785 | No |
| 55 | NDUFV1 | NDUFV1 Entrez, Source | NADH dehydrogenase (ubiquinone) flavoprotein 1, 51kDa | 5933 | 0.001 | 0.2562 | No |
| 56 | SH3BP4 | SH3BP4 Entrez, Source | SH3-domain binding protein 4 | 6416 | -0.007 | 0.2203 | No |
| 57 | CNR1 | CNR1 Entrez, Source | cannabinoid receptor 1 (brain) | 6660 | -0.010 | 0.2028 | No |
| 58 | GSTM5 | GSTM5 Entrez, Source | glutathione S-transferase M5 | 6680 | -0.010 | 0.2021 | No |
| 59 | CBR1 | CBR1 Entrez, Source | carbonyl reductase 1 | 7273 | -0.019 | 0.1589 | No |
| 60 | GATA2 | GATA2 Entrez, Source | GATA binding protein 2 | 7313 | -0.020 | 0.1575 | No |
| 61 | DMN | DMN Entrez, Source | desmuslin | 7569 | -0.024 | 0.1401 | No |
| 62 | HIF1A | HIF1A Entrez, Source | hypoxia-inducible factor 1, alpha subunit (basic helix-loop-helix transcription factor) | 7751 | -0.027 | 0.1285 | No |
| 63 | DDX5 | DDX5 Entrez, Source | DEAD (Asp-Glu-Ala-Asp) box polypeptide 5 | 8037 | -0.031 | 0.1094 | No |
| 64 | MAPK6 | MAPK6 Entrez, Source | mitogen-activated protein kinase 6 | 8336 | -0.036 | 0.0897 | No |
| 65 | ITGB4BP | ITGB4BP Entrez, Source | integrin beta 4 binding protein | 8464 | -0.038 | 0.0831 | No |
| 66 | SDC1 | SDC1 Entrez, Source | syndecan 1 | 8872 | -0.045 | 0.0558 | No |
| 67 | PTGS2 | PTGS2 Entrez, Source | prostaglandin-endoperoxide synthase 2 (prostaglandin G/H synthase and cyclooxygenase) | 8937 | -0.046 | 0.0546 | No |
| 68 | GSK3B | GSK3B Entrez, Source | glycogen synthase kinase 3 beta | 8958 | -0.047 | 0.0567 | No |
| 69 | PDE8B | PDE8B Entrez, Source | phosphodiesterase 8B | 9248 | -0.052 | 0.0389 | No |
| 70 | SYNJ2 | SYNJ2 Entrez, Source | synaptojanin 2 | 9325 | -0.053 | 0.0372 | No |
| 71 | GFAP | GFAP Entrez, Source | glial fibrillary acidic protein | 9399 | -0.054 | 0.0359 | No |
| 72 | PHB | PHB Entrez, Source | prohibitin | 9567 | -0.057 | 0.0278 | No |
| 73 | GNAS | GNAS Entrez, Source | GNAS complex locus | 9688 | -0.060 | 0.0233 | No |
| 74 | NME2 | NME2 Entrez, Source | non-metastatic cells 2, protein (NM23B) expressed in | 9773 | -0.061 | 0.0217 | No |
| 75 | HSPE1 | HSPE1 Entrez, Source | heat shock 10kDa protein 1 (chaperonin 10) | 9837 | -0.063 | 0.0219 | No |
| 76 | DDX3X | DDX3X Entrez, Source | DEAD (Asp-Glu-Ala-Asp) box polypeptide 3, X-linked | 9904 | -0.064 | 0.0219 | No |
| 77 | RPL32 | RPL32 Entrez, Source | ribosomal protein L32 | 10048 | -0.067 | 0.0163 | No |
| 78 | RPL8 | RPL8 Entrez, Source | ribosomal protein L8 | 10123 | -0.069 | 0.0160 | No |
| 79 | MRPL2 | MRPL2 Entrez, Source | mitochondrial ribosomal protein L2 | 10203 | -0.071 | 0.0155 | No |
| 80 | TARS | TARS Entrez, Source | threonyl-tRNA synthetase | 10249 | -0.072 | 0.0177 | No |
| 81 | METAP2 | METAP2 Entrez, Source | methionyl aminopeptidase 2 | 10345 | -0.074 | 0.0162 | No |
| 82 | ATP6V1C1 | ATP6V1C1 Entrez, Source | ATPase, H+ transporting, lysosomal 42kDa, V1 subunit C1 | 10713 | -0.083 | -0.0050 | No |
| 83 | EIF3S6 | EIF3S6 Entrez, Source | eukaryotic translation initiation factor 3, subunit 6 48kDa | 10822 | -0.086 | -0.0065 | No |
| 84 | BHLHB2 | BHLHB2 Entrez, Source | basic helix-loop-helix domain containing, class B, 2 | 10960 | -0.089 | -0.0099 | No |
| 85 | TKT | TKT Entrez, Source | transketolase (Wernicke-Korsakoff syndrome) | 11381 | -0.101 | -0.0338 | No |
| 86 | VEGF | VEGF Entrez, Source | vascular endothelial growth factor | 11386 | -0.102 | -0.0262 | No |
| 87 | IRF1 | IRF1 Entrez, Source | interferon regulatory factor 1 | 11466 | -0.105 | -0.0241 | No |
| 88 | PTEN | PTEN Entrez, Source | phosphatase and tensin homolog (mutated in multiple advanced cancers 1) | 11578 | -0.108 | -0.0240 | No |
| 89 | EEF1D | EEF1D Entrez, Source | eukaryotic translation elongation factor 1 delta (guanine nucleotide exchange protein) | 11614 | -0.110 | -0.0182 | No |
| 90 | STIP1 | STIP1 Entrez, Source | stress-induced-phosphoprotein 1 (Hsp70/Hsp90-organizing protein) | 11703 | -0.113 | -0.0160 | No |
| 91 | VLDLR | VLDLR Entrez, Source | very low density lipoprotein receptor | 12037 | -0.128 | -0.0312 | No |
| 92 | NUP93 | NUP93 Entrez, Source | nucleoporin 93kDa | 12085 | -0.131 | -0.0246 | No |
| 93 | SF3A3 | SF3A3 Entrez, Source | splicing factor 3a, subunit 3, 60kDa | 12128 | -0.133 | -0.0175 | No |
| 94 | DDX1 | DDX1 Entrez, Source | DEAD (Asp-Glu-Ala-Asp) box polypeptide 1 | 12162 | -0.135 | -0.0095 | No |
| 95 | PPIE | PPIE Entrez, Source | peptidylprolyl isomerase E (cyclophilin E) | 12220 | -0.137 | -0.0031 | No |
| 96 | CD28 | CD28 Entrez, Source | CD28 molecule | 12234 | -0.138 | 0.0066 | No |
| 97 | KLF10 | KLF10 Entrez, Source | Kruppel-like factor 10 | 12453 | -0.151 | 0.0019 | No |
| 98 | LY6G6C | LY6G6C Entrez, Source | lymphocyte antigen 6 complex, locus G6C | 12486 | -0.154 | 0.0114 | No |
| 99 | THOC1 | THOC1 Entrez, Source | THO complex 1 | 12520 | -0.156 | 0.0210 | No |
| 100 | UGP2 | UGP2 Entrez, Source | UDP-glucose pyrophosphorylase 2 | 12627 | -0.166 | 0.0259 | No |
| 101 | KHK | KHK Entrez, Source | ketohexokinase (fructokinase) | 12757 | -0.178 | 0.0299 | No |
| 102 | GPX1 | GPX1 Entrez, Source | glutathione peroxidase 1 | 12783 | -0.183 | 0.0422 | No |