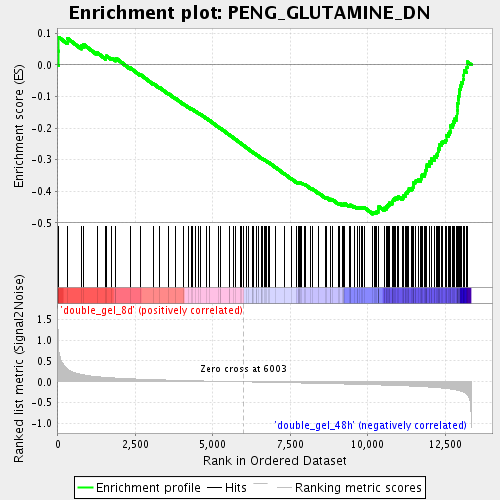
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | double_gel_and_single_gel_rma_expression_values_collapsed_to_symbols.class.cls #double_gel_8d_versus_double_gel_48h.class.cls #double_gel_8d_versus_double_gel_48h_repos |
| Phenotype | class.cls#double_gel_8d_versus_double_gel_48h_repos |
| Upregulated in class | double_gel_48h |
| GeneSet | PENG_GLUTAMINE_DN |
| Enrichment Score (ES) | -0.47235242 |
| Normalized Enrichment Score (NES) | -2.0157437 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.0075870575 |
| FWER p-Value | 0.101 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | LDHB | LDHB Entrez, Source | lactate dehydrogenase B | 22 | 0.785 | 0.0442 | No |
| 2 | PFKP | PFKP Entrez, Source | phosphofructokinase, platelet | 29 | 0.753 | 0.0877 | No |
| 3 | HSPA4 | HSPA4 Entrez, Source | heat shock 70kDa protein 4 | 305 | 0.303 | 0.0845 | No |
| 4 | EMP3 | EMP3 Entrez, Source | epithelial membrane protein 3 | 755 | 0.176 | 0.0607 | No |
| 5 | TGFB1 | TGFB1 Entrez, Source | transforming growth factor, beta 1 (Camurati-Engelmann disease) | 821 | 0.166 | 0.0654 | No |
| 6 | CLIC1 | CLIC1 Entrez, Source | chloride intracellular channel 1 | 1263 | 0.123 | 0.0391 | No |
| 7 | PSMD1 | PSMD1 Entrez, Source | proteasome (prosome, macropain) 26S subunit, non-ATPase, 1 | 1543 | 0.105 | 0.0240 | No |
| 8 | ALDOA | ALDOA Entrez, Source | aldolase A, fructose-bisphosphate | 1564 | 0.104 | 0.0286 | No |
| 9 | IDH3A | IDH3A Entrez, Source | isocitrate dehydrogenase 3 (NAD+) alpha | 1728 | 0.096 | 0.0219 | No |
| 10 | SCD | SCD Entrez, Source | stearoyl-CoA desaturase (delta-9-desaturase) | 1854 | 0.090 | 0.0176 | No |
| 11 | GJB1 | GJB1 Entrez, Source | gap junction protein, beta 1, 32kDa (connexin 32, Charcot-Marie-Tooth neuropathy, X-linked) | 1871 | 0.090 | 0.0217 | No |
| 12 | MYD88 | MYD88 Entrez, Source | myeloid differentiation primary response gene (88) | 2332 | 0.073 | -0.0090 | No |
| 13 | CCND1 | CCND1 Entrez, Source | cyclin D1 | 2668 | 0.063 | -0.0308 | No |
| 14 | SQLE | SQLE Entrez, Source | squalene epoxidase | 3091 | 0.052 | -0.0598 | No |
| 15 | PSMA6 | PSMA6 Entrez, Source | proteasome (prosome, macropain) subunit, alpha type, 6 | 3279 | 0.048 | -0.0712 | No |
| 16 | MAP3K11 | MAP3K11 Entrez, Source | mitogen-activated protein kinase kinase kinase 11 | 3563 | 0.042 | -0.0903 | No |
| 17 | PSMD8 | PSMD8 Entrez, Source | proteasome (prosome, macropain) 26S subunit, non-ATPase, 8 | 3796 | 0.037 | -0.1058 | No |
| 18 | HRAS | HRAS Entrez, Source | v-Ha-ras Harvey rat sarcoma viral oncogene homolog | 4055 | 0.032 | -0.1235 | No |
| 19 | SYNGR2 | SYNGR2 Entrez, Source | synaptogyrin 2 | 4223 | 0.029 | -0.1345 | No |
| 20 | IFI30 | IFI30 Entrez, Source | interferon, gamma-inducible protein 30 | 4301 | 0.027 | -0.1388 | No |
| 21 | PPIF | PPIF Entrez, Source | peptidylprolyl isomerase F (cyclophilin F) | 4337 | 0.027 | -0.1399 | No |
| 22 | ADRM1 | ADRM1 Entrez, Source | adhesion regulating molecule 1 | 4443 | 0.025 | -0.1464 | No |
| 23 | ODC1 | ODC1 Entrez, Source | ornithine decarboxylase 1 | 4552 | 0.022 | -0.1533 | No |
| 24 | CELSR2 | CELSR2 Entrez, Source | cadherin, EGF LAG seven-pass G-type receptor 2 (flamingo homolog, Drosophila) | 4608 | 0.021 | -0.1563 | No |
| 25 | OGFR | OGFR Entrez, Source | opioid growth factor receptor | 4793 | 0.019 | -0.1692 | No |
| 26 | CYCS | CYCS Entrez, Source | cytochrome c, somatic | 4888 | 0.017 | -0.1753 | No |
| 27 | IDH1 | IDH1 Entrez, Source | isocitrate dehydrogenase 1 (NADP+), soluble | 5182 | 0.012 | -0.1969 | No |
| 28 | PIK3R2 | PIK3R2 Entrez, Source | phosphoinositide-3-kinase, regulatory subunit 2 (p85 beta) | 5237 | 0.011 | -0.2003 | No |
| 29 | PPIB | PPIB Entrez, Source | peptidylprolyl isomerase B (cyclophilin B) | 5253 | 0.011 | -0.2008 | No |
| 30 | LDHA | LDHA Entrez, Source | lactate dehydrogenase A | 5536 | 0.007 | -0.2219 | No |
| 31 | TUBG1 | TUBG1 Entrez, Source | tubulin, gamma 1 | 5540 | 0.007 | -0.2217 | No |
| 32 | STAT1 | STAT1 Entrez, Source | signal transducer and activator of transcription 1, 91kDa | 5682 | 0.005 | -0.2321 | No |
| 33 | LIMK2 | LIMK2 Entrez, Source | LIM domain kinase 2 | 5743 | 0.004 | -0.2365 | No |
| 34 | NMT1 | NMT1 Entrez, Source | N-myristoyltransferase 1 | 5907 | 0.001 | -0.2488 | No |
| 35 | ATP5G3 | ATP5G3 Entrez, Source | ATP synthase, H+ transporting, mitochondrial F0 complex, subunit C3 (subunit 9) | 5923 | 0.001 | -0.2499 | No |
| 36 | CDC25A | CDC25A Entrez, Source | cell division cycle 25A | 6002 | 0.000 | -0.2558 | No |
| 37 | TOP1 | TOP1 Entrez, Source | topoisomerase (DNA) I | 6078 | -0.001 | -0.2614 | No |
| 38 | NDUFS6 | NDUFS6 Entrez, Source | NADH dehydrogenase (ubiquinone) Fe-S protein 6, 13kDa (NADH-coenzyme Q reductase) | 6136 | -0.002 | -0.2656 | No |
| 39 | ADAM19 | ADAM19 Entrez, Source | ADAM metallopeptidase domain 19 (meltrin beta) | 6281 | -0.004 | -0.2763 | No |
| 40 | COPS3 | COPS3 Entrez, Source | COP9 constitutive photomorphogenic homolog subunit 3 (Arabidopsis) | 6295 | -0.005 | -0.2770 | No |
| 41 | SF3B4 | SF3B4 Entrez, Source | splicing factor 3b, subunit 4, 49kDa | 6322 | -0.005 | -0.2787 | No |
| 42 | PTMS | PTMS Entrez, Source | parathymosin | 6405 | -0.006 | -0.2846 | No |
| 43 | GCSH | GCSH Entrez, Source | glycine cleavage system protein H (aminomethyl carrier) | 6406 | -0.006 | -0.2842 | No |
| 44 | GRIN2A | GRIN2A Entrez, Source | glutamate receptor, ionotropic, N-methyl D-aspartate 2A | 6460 | -0.007 | -0.2878 | No |
| 45 | NUP98 | NUP98 Entrez, Source | nucleoporin 98kDa | 6561 | -0.008 | -0.2949 | No |
| 46 | PSMD12 | PSMD12 Entrez, Source | proteasome (prosome, macropain) 26S subunit, non-ATPase, 12 | 6575 | -0.009 | -0.2954 | No |
| 47 | HSPA8 | HSPA8 Entrez, Source | heat shock 70kDa protein 8 | 6613 | -0.009 | -0.2977 | No |
| 48 | SLC19A1 | SLC19A1 Entrez, Source | solute carrier family 19 (folate transporter), member 1 | 6656 | -0.010 | -0.3003 | No |
| 49 | PPP2R2A | PPP2R2A Entrez, Source | protein phosphatase 2 (formerly 2A), regulatory subunit B (PR 52), alpha isoform | 6690 | -0.011 | -0.3022 | No |
| 50 | DNAJB6 | DNAJB6 Entrez, Source | DnaJ (Hsp40) homolog, subfamily B, member 6 | 6729 | -0.011 | -0.3044 | No |
| 51 | CCNB1 | CCNB1 Entrez, Source | cyclin B1 | 6785 | -0.012 | -0.3079 | No |
| 52 | BCAP31 | BCAP31 Entrez, Source | B-cell receptor-associated protein 31 | 6822 | -0.012 | -0.3099 | No |
| 53 | VCP | VCP Entrez, Source | valosin-containing protein | 7037 | -0.016 | -0.3252 | No |
| 54 | POLR2C | POLR2C Entrez, Source | polymerase (RNA) II (DNA directed) polypeptide C, 33kDa | 7301 | -0.020 | -0.3441 | No |
| 55 | UBE2N | UBE2N Entrez, Source | ubiquitin-conjugating enzyme E2N (UBC13 homolog, yeast) | 7534 | -0.023 | -0.3603 | No |
| 56 | PDCD4 | PDCD4 Entrez, Source | programmed cell death 4 (neoplastic transformation inhibitor) | 7707 | -0.026 | -0.3719 | No |
| 57 | DHCR24 | DHCR24 Entrez, Source | 24-dehydrocholesterol reductase | 7755 | -0.027 | -0.3739 | No |
| 58 | ACLY | ACLY Entrez, Source | ATP citrate lyase | 7774 | -0.027 | -0.3737 | No |
| 59 | EIF2B4 | EIF2B4 Entrez, Source | eukaryotic translation initiation factor 2B, subunit 4 delta, 67kDa | 7792 | -0.027 | -0.3734 | No |
| 60 | KPNA2 | KPNA2 Entrez, Source | karyopherin alpha 2 (RAG cohort 1, importin alpha 1) | 7811 | -0.028 | -0.3732 | No |
| 61 | PSMD3 | PSMD3 Entrez, Source | proteasome (prosome, macropain) 26S subunit, non-ATPase, 3 | 7835 | -0.028 | -0.3733 | No |
| 62 | PGAM1 | PGAM1 Entrez, Source | phosphoglycerate mutase 1 (brain) | 7875 | -0.029 | -0.3746 | No |
| 63 | ATP2A2 | ATP2A2 Entrez, Source | ATPase, Ca++ transporting, cardiac muscle, slow twitch 2 | 7943 | -0.030 | -0.3779 | No |
| 64 | VDAC1 | VDAC1 Entrez, Source | voltage-dependent anion channel 1 | 7979 | -0.030 | -0.3788 | No |
| 65 | PIK3R3 | PIK3R3 Entrez, Source | phosphoinositide-3-kinase, regulatory subunit 3 (p55, gamma) | 8161 | -0.033 | -0.3906 | No |
| 66 | DNAJA1 | DNAJA1 Entrez, Source | DnaJ (Hsp40) homolog, subfamily A, member 1 | 8213 | -0.034 | -0.3925 | No |
| 67 | TCEB1 | TCEB1 Entrez, Source | transcription elongation factor B (SIII), polypeptide 1 (15kDa, elongin C) | 8413 | -0.038 | -0.4054 | No |
| 68 | PRDX4 | PRDX4 Entrez, Source | peroxiredoxin 4 | 8646 | -0.041 | -0.4206 | No |
| 69 | TXNIP | TXNIP Entrez, Source | thioredoxin interacting protein | 8667 | -0.041 | -0.4197 | No |
| 70 | HPRT1 | HPRT1 Entrez, Source | hypoxanthine phosphoribosyltransferase 1 (Lesch-Nyhan syndrome) | 8796 | -0.044 | -0.4269 | No |
| 71 | G3BP | G3BP Entrez, Source | - | 8799 | -0.044 | -0.4245 | No |
| 72 | RNASEH1 | RNASEH1 Entrez, Source | ribonuclease H1 | 8861 | -0.045 | -0.4265 | No |
| 73 | CALR | CALR Entrez, Source | calreticulin | 9052 | -0.048 | -0.4382 | No |
| 74 | MORF4L2 | MORF4L2 Entrez, Source | mortality factor 4 like 2 | 9092 | -0.049 | -0.4383 | No |
| 75 | PRODH | PRODH Entrez, Source | proline dehydrogenase (oxidase) 1 | 9179 | -0.050 | -0.4419 | No |
| 76 | PSMB3 | PSMB3 Entrez, Source | proteasome (prosome, macropain) subunit, beta type, 3 | 9221 | -0.051 | -0.4420 | No |
| 77 | ATP1B3 | ATP1B3 Entrez, Source | ATPase, Na+/K+ transporting, beta 3 polypeptide | 9230 | -0.051 | -0.4396 | No |
| 78 | PPP1R10 | PPP1R10 Entrez, Source | protein phosphatase 1, regulatory subunit 10 | 9241 | -0.051 | -0.4374 | No |
| 79 | XRCC5 | XRCC5 Entrez, Source | X-ray repair complementing defective repair in Chinese hamster cells 5 (double-strand-break rejoining; Ku autoantigen, 80kDa) | 9405 | -0.054 | -0.4466 | No |
| 80 | CTSC | CTSC Entrez, Source | cathepsin C | 9429 | -0.055 | -0.4452 | No |
| 81 | SGTA | SGTA Entrez, Source | small glutamine-rich tetratricopeptide repeat (TPR)-containing, alpha | 9449 | -0.055 | -0.4434 | No |
| 82 | PHB | PHB Entrez, Source | prohibitin | 9567 | -0.057 | -0.4489 | No |
| 83 | EIF5 | EIF5 Entrez, Source | eukaryotic translation initiation factor 5 | 9653 | -0.059 | -0.4520 | No |
| 84 | GMPS | GMPS Entrez, Source | guanine monphosphate synthetase | 9684 | -0.060 | -0.4508 | No |
| 85 | EBP | EBP Entrez, Source | emopamil binding protein (sterol isomerase) | 9744 | -0.061 | -0.4517 | No |
| 86 | EIF4G2 | EIF4G2 Entrez, Source | eukaryotic translation initiation factor 4 gamma, 2 | 9796 | -0.062 | -0.4519 | No |
| 87 | HSPE1 | HSPE1 Entrez, Source | heat shock 10kDa protein 1 (chaperonin 10) | 9837 | -0.063 | -0.4513 | No |
| 88 | MRPL23 | MRPL23 Entrez, Source | mitochondrial ribosomal protein L23 | 9879 | -0.064 | -0.4507 | No |
| 89 | PTPN6 | PTPN6 Entrez, Source | protein tyrosine phosphatase, non-receptor type 6 | 10165 | -0.070 | -0.4683 | Yes |
| 90 | TCP1 | TCP1 Entrez, Source | t-complex 1 | 10212 | -0.071 | -0.4677 | Yes |
| 91 | CCT6A | CCT6A Entrez, Source | chaperonin containing TCP1, subunit 6A (zeta 1) | 10250 | -0.072 | -0.4663 | Yes |
| 92 | PRDX1 | PRDX1 Entrez, Source | peroxiredoxin 1 | 10293 | -0.072 | -0.4653 | Yes |
| 93 | MRPL12 | MRPL12 Entrez, Source | mitochondrial ribosomal protein L12 | 10333 | -0.073 | -0.4639 | Yes |
| 94 | CCT2 | CCT2 Entrez, Source | chaperonin containing TCP1, subunit 2 (beta) | 10334 | -0.073 | -0.4596 | Yes |
| 95 | RBBP6 | RBBP6 Entrez, Source | retinoblastoma binding protein 6 | 10335 | -0.073 | -0.4554 | Yes |
| 96 | AK2 | AK2 Entrez, Source | adenylate kinase 2 | 10352 | -0.074 | -0.4523 | Yes |
| 97 | PRPF4 | PRPF4 Entrez, Source | PRP4 pre-mRNA processing factor 4 homolog (yeast) | 10359 | -0.074 | -0.4484 | Yes |
| 98 | GSPT1 | GSPT1 Entrez, Source | G1 to S phase transition 1 | 10533 | -0.079 | -0.4570 | Yes |
| 99 | FKBP4 | FKBP4 Entrez, Source | FK506 binding protein 4, 59kDa | 10541 | -0.079 | -0.4529 | Yes |
| 100 | RABGGTB | RABGGTB Entrez, Source | Rab geranylgeranyltransferase, beta subunit | 10595 | -0.080 | -0.4523 | Yes |
| 101 | RANGAP1 | RANGAP1 Entrez, Source | Ran GTPase activating protein 1 | 10603 | -0.080 | -0.4482 | Yes |
| 102 | TIMM17A | TIMM17A Entrez, Source | translocase of inner mitochondrial membrane 17 homolog A (yeast) | 10631 | -0.081 | -0.4455 | Yes |
| 103 | ADCY3 | ADCY3 Entrez, Source | adenylate cyclase 3 | 10657 | -0.081 | -0.4426 | Yes |
| 104 | SFRS2 | SFRS2 Entrez, Source | splicing factor, arginine/serine-rich 2 | 10691 | -0.082 | -0.4403 | Yes |
| 105 | NUDC | NUDC Entrez, Source | nuclear distribution gene C homolog (A. nidulans) | 10710 | -0.083 | -0.4369 | Yes |
| 106 | UMPS | UMPS Entrez, Source | uridine monophosphate synthetase (orotate phosphoribosyl transferase and orotidine-5'-decarboxylase) | 10783 | -0.085 | -0.4374 | Yes |
| 107 | PSME4 | PSME4 Entrez, Source | proteasome (prosome, macropain) activator subunit 4 | 10789 | -0.085 | -0.4328 | Yes |
| 108 | RPS26 | RPS26 Entrez, Source | ribosomal protein S26 | 10813 | -0.086 | -0.4296 | Yes |
| 109 | PFN1 | PFN1 Entrez, Source | profilin 1 | 10821 | -0.086 | -0.4251 | Yes |
| 110 | PABPN1 | PABPN1 Entrez, Source | poly(A) binding protein, nuclear 1 | 10867 | -0.087 | -0.4234 | Yes |
| 111 | RNF126 | RNF126 Entrez, Source | ring finger protein 126 | 10907 | -0.088 | -0.4212 | Yes |
| 112 | HSPA5 | HSPA5 Entrez, Source | heat shock 70kDa protein 5 (glucose-regulated protein, 78kDa) | 10956 | -0.089 | -0.4196 | Yes |
| 113 | C1QBP | C1QBP Entrez, Source | complement component 1, q subcomponent binding protein | 10991 | -0.090 | -0.4169 | Yes |
| 114 | BZW1 | BZW1 Entrez, Source | basic leucine zipper and W2 domains 1 | 11133 | -0.094 | -0.4222 | Yes |
| 115 | DHCR7 | DHCR7 Entrez, Source | 7-dehydrocholesterol reductase | 11141 | -0.094 | -0.4172 | Yes |
| 116 | HNRPL | HNRPL Entrez, Source | heterogeneous nuclear ribonucleoprotein L | 11160 | -0.095 | -0.4130 | Yes |
| 117 | MTHFD1 | MTHFD1 Entrez, Source | methylenetetrahydrofolate dehydrogenase (NADP+ dependent) 1, methenyltetrahydrofolate cyclohydrolase, formyltetrahydrofolate synthetase | 11221 | -0.096 | -0.4120 | Yes |
| 118 | DKC1 | DKC1 Entrez, Source | dyskeratosis congenita 1, dyskerin | 11229 | -0.096 | -0.4069 | Yes |
| 119 | EWSR1 | EWSR1 Entrez, Source | Ewing sarcoma breakpoint region 1 | 11248 | -0.097 | -0.4026 | Yes |
| 120 | EIF5A | EIF5A Entrez, Source | eukaryotic translation initiation factor 5A | 11294 | -0.098 | -0.4002 | Yes |
| 121 | PTDSS1 | PTDSS1 Entrez, Source | phosphatidylserine synthase 1 | 11318 | -0.099 | -0.3962 | Yes |
| 122 | LMNB1 | LMNB1 Entrez, Source | lamin B1 | 11330 | -0.100 | -0.3912 | Yes |
| 123 | HNRPAB | HNRPAB Entrez, Source | heterogeneous nuclear ribonucleoprotein A/B | 11401 | -0.102 | -0.3906 | Yes |
| 124 | TBL3 | TBL3 Entrez, Source | transducin (beta)-like 3 | 11459 | -0.105 | -0.3888 | Yes |
| 125 | ABCF1 | ABCF1 Entrez, Source | ATP-binding cassette, sub-family F (GCN20), member 1 | 11469 | -0.105 | -0.3833 | Yes |
| 126 | CCT5 | CCT5 Entrez, Source | chaperonin containing TCP1, subunit 5 (epsilon) | 11472 | -0.105 | -0.3774 | Yes |
| 127 | PSMB6 | PSMB6 Entrez, Source | proteasome (prosome, macropain) subunit, beta type, 6 | 11483 | -0.105 | -0.3720 | Yes |
| 128 | KARS | KARS Entrez, Source | lysyl-tRNA synthetase | 11525 | -0.107 | -0.3689 | Yes |
| 129 | BMS1L | BMS1L Entrez, Source | BMS1-like, ribosome assembly protein (yeast) | 11556 | -0.108 | -0.3649 | Yes |
| 130 | SLC5A6 | SLC5A6 Entrez, Source | solute carrier family 5 (sodium-dependent vitamin transporter), member 6 | 11622 | -0.110 | -0.3634 | Yes |
| 131 | BYSL | BYSL Entrez, Source | bystin-like | 11704 | -0.113 | -0.3629 | Yes |
| 132 | TOMM40 | TOMM40 Entrez, Source | translocase of outer mitochondrial membrane 40 homolog (yeast) | 11720 | -0.114 | -0.3574 | Yes |
| 133 | PAICS | PAICS Entrez, Source | phosphoribosylaminoimidazole carboxylase, phosphoribosylaminoimidazole succinocarboxamide synthetase | 11730 | -0.114 | -0.3514 | Yes |
| 134 | BOP1 | BOP1 Entrez, Source | block of proliferation 1 | 11761 | -0.115 | -0.3469 | Yes |
| 135 | PRPS1 | PRPS1 Entrez, Source | phosphoribosyl pyrophosphate synthetase 1 | 11832 | -0.118 | -0.3454 | Yes |
| 136 | PRKCBP1 | PRKCBP1 Entrez, Source | protein kinase C binding protein 1 | 11848 | -0.119 | -0.3395 | Yes |
| 137 | SSBP1 | SSBP1 Entrez, Source | single-stranded DNA binding protein 1 | 11852 | -0.119 | -0.3328 | Yes |
| 138 | FDFT1 | FDFT1 Entrez, Source | farnesyl-diphosphate farnesyltransferase 1 | 11893 | -0.121 | -0.3288 | Yes |
| 139 | DDX18 | DDX18 Entrez, Source | DEAD (Asp-Glu-Ala-Asp) box polypeptide 18 | 11900 | -0.122 | -0.3221 | Yes |
| 140 | EIF4G1 | EIF4G1 Entrez, Source | eukaryotic translation initiation factor 4 gamma, 1 | 11911 | -0.122 | -0.3158 | Yes |
| 141 | SC4MOL | SC4MOL Entrez, Source | sterol-C4-methyl oxidase-like | 11986 | -0.126 | -0.3140 | Yes |
| 142 | SSB | SSB Entrez, Source | Sjogren syndrome antigen B (autoantigen La) | 11991 | -0.126 | -0.3070 | Yes |
| 143 | CHD4 | CHD4 Entrez, Source | chromodomain helicase DNA binding protein 4 | 12051 | -0.129 | -0.3039 | Yes |
| 144 | FASN | FASN Entrez, Source | fatty acid synthase | 12064 | -0.130 | -0.2973 | Yes |
| 145 | XBP1 | XBP1 Entrez, Source | X-box binding protein 1 | 12147 | -0.134 | -0.2957 | Yes |
| 146 | DDX1 | DDX1 Entrez, Source | DEAD (Asp-Glu-Ala-Asp) box polypeptide 1 | 12162 | -0.135 | -0.2889 | Yes |
| 147 | CDC20 | CDC20 Entrez, Source | CDC20 cell division cycle 20 homolog (S. cerevisiae) | 12218 | -0.137 | -0.2850 | Yes |
| 148 | ALG8 | ALG8 Entrez, Source | asparagine-linked glycosylation 8 homolog (S. cerevisiae, alpha-1,3-glucosyltransferase) | 12261 | -0.140 | -0.2801 | Yes |
| 149 | KHSRP | KHSRP Entrez, Source | KH-type splicing regulatory protein (FUSE binding protein 2) | 12267 | -0.140 | -0.2722 | Yes |
| 150 | TARDBP | TARDBP Entrez, Source | TAR DNA binding protein | 12292 | -0.142 | -0.2658 | Yes |
| 151 | RUVBL2 | RUVBL2 Entrez, Source | RuvB-like 2 (E. coli) | 12316 | -0.144 | -0.2591 | Yes |
| 152 | NME1 | NME1 Entrez, Source | non-metastatic cells 1, protein (NM23A) expressed in | 12324 | -0.144 | -0.2512 | Yes |
| 153 | AMD1 | AMD1 Entrez, Source | adenosylmethionine decarboxylase 1 | 12376 | -0.147 | -0.2465 | Yes |
| 154 | SRPK1 | SRPK1 Entrez, Source | SFRS protein kinase 1 | 12424 | -0.150 | -0.2413 | Yes |
| 155 | NOL5A | NOL5A Entrez, Source | nucleolar protein 5A (56kDa with KKE/D repeat) | 12500 | -0.155 | -0.2380 | Yes |
| 156 | IDI1 | IDI1 Entrez, Source | isopentenyl-diphosphate delta isomerase 1 | 12532 | -0.157 | -0.2312 | Yes |
| 157 | RQCD1 | RQCD1 Entrez, Source | RCD1 required for cell differentiation1 homolog (S. pombe) | 12533 | -0.157 | -0.2220 | Yes |
| 158 | LAMB3 | LAMB3 Entrez, Source | laminin, beta 3 | 12600 | -0.163 | -0.2175 | Yes |
| 159 | ALG3 | ALG3 Entrez, Source | asparagine-linked glycosylation 3 homolog (S. cerevisiae, alpha-1,3-mannosyltransferase) | 12651 | -0.168 | -0.2115 | Yes |
| 160 | EIF2B5 | EIF2B5 Entrez, Source | eukaryotic translation initiation factor 2B, subunit 5 epsilon, 82kDa | 12669 | -0.170 | -0.2028 | Yes |
| 161 | TRIP13 | TRIP13 Entrez, Source | thyroid hormone receptor interactor 13 | 12670 | -0.170 | -0.1929 | Yes |
| 162 | SYNCRIP | SYNCRIP Entrez, Source | synaptotagmin binding, cytoplasmic RNA interacting protein | 12730 | -0.176 | -0.1871 | Yes |
| 163 | DYRK1A | DYRK1A Entrez, Source | dual-specificity tyrosine-(Y)-phosphorylation regulated kinase 1A | 12773 | -0.181 | -0.1797 | Yes |
| 164 | EBNA1BP2 | EBNA1BP2 Entrez, Source | EBNA1 binding protein 2 | 12803 | -0.186 | -0.1711 | Yes |
| 165 | TAP1 | TAP1 Entrez, Source | transporter 1, ATP-binding cassette, sub-family B (MDR/TAP) | 12854 | -0.192 | -0.1637 | Yes |
| 166 | FBL | FBL Entrez, Source | fibrillarin | 12880 | -0.197 | -0.1540 | Yes |
| 167 | SRM | SRM Entrez, Source | spermidine synthase | 12882 | -0.197 | -0.1426 | Yes |
| 168 | TDG | TDG Entrez, Source | thymine-DNA glycosylase | 12893 | -0.199 | -0.1318 | Yes |
| 169 | SLC29A2 | SLC29A2 Entrez, Source | solute carrier family 29 (nucleoside transporters), member 2 | 12900 | -0.200 | -0.1206 | Yes |
| 170 | NCL | NCL Entrez, Source | nucleolin | 12916 | -0.202 | -0.1099 | Yes |
| 171 | SLC29A1 | SLC29A1 Entrez, Source | solute carrier family 29 (nucleoside transporters), member 1 | 12934 | -0.205 | -0.0992 | Yes |
| 172 | SLC4A7 | SLC4A7 Entrez, Source | solute carrier family 4, sodium bicarbonate cotransporter, member 7 | 12948 | -0.209 | -0.0880 | Yes |
| 173 | ACAT2 | ACAT2 Entrez, Source | acetyl-Coenzyme A acetyltransferase 2 (acetoacetyl Coenzyme A thiolase) | 12971 | -0.213 | -0.0772 | Yes |
| 174 | XPO1 | XPO1 Entrez, Source | exportin 1 (CRM1 homolog, yeast) | 12994 | -0.216 | -0.0663 | Yes |
| 175 | NOLC1 | NOLC1 Entrez, Source | nucleolar and coiled-body phosphoprotein 1 | 13015 | -0.221 | -0.0548 | Yes |
| 176 | PA2G4 | PA2G4 Entrez, Source | proliferation-associated 2G4, 38kDa | 13074 | -0.242 | -0.0451 | Yes |
| 177 | FUS | FUS Entrez, Source | fusion (involved in t(12;16) in malignant liposarcoma) | 13100 | -0.252 | -0.0323 | Yes |
| 178 | INHBC | INHBC Entrez, Source | inhibin, beta C | 13106 | -0.255 | -0.0178 | Yes |
| 179 | TFRC | TFRC Entrez, Source | transferrin receptor (p90, CD71) | 13197 | -0.299 | -0.0072 | Yes |
| 180 | SFPQ | SFPQ Entrez, Source | splicing factor proline/glutamine-rich (polypyrimidine tract binding protein associated) | 13213 | -0.310 | 0.0098 | Yes |