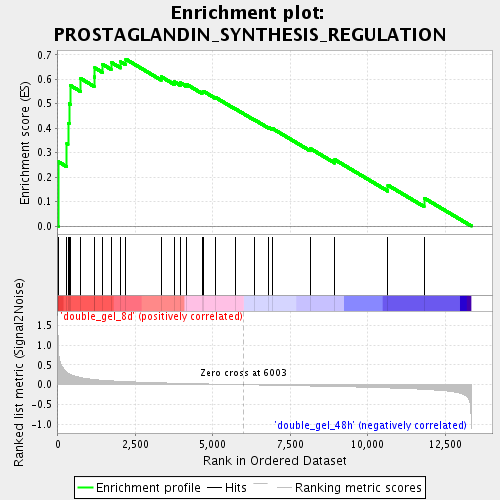
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | double_gel_and_single_gel_rma_expression_values_collapsed_to_symbols.class.cls #double_gel_8d_versus_double_gel_48h.class.cls #double_gel_8d_versus_double_gel_48h_repos |
| Phenotype | class.cls#double_gel_8d_versus_double_gel_48h_repos |
| Upregulated in class | double_gel_8d |
| GeneSet | PROSTAGLANDIN_SYNTHESIS_REGULATION |
| Enrichment Score (ES) | 0.682141 |
| Normalized Enrichment Score (NES) | 1.9327979 |
| Nominal p-value | 0.0037807184 |
| FDR q-value | 0.008831204 |
| FWER p-Value | 0.457 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | ANXA1 | ANXA1 Entrez, Source | annexin A1 | 11 | 0.884 | 0.2632 | Yes |
| 2 | ANXA8 | ANXA8 Entrez, Source | annexin A8 | 289 | 0.316 | 0.3368 | Yes |
| 3 | EDN1 | EDN1 Entrez, Source | endothelin 1 | 326 | 0.291 | 0.4211 | Yes |
| 4 | S100A6 | S100A6 Entrez, Source | S100 calcium binding protein A6 | 365 | 0.274 | 0.5001 | Yes |
| 5 | PTGER4 | PTGER4 Entrez, Source | prostaglandin E receptor 4 (subtype EP4) | 402 | 0.258 | 0.5746 | Yes |
| 6 | ANXA4 | ANXA4 Entrez, Source | annexin A4 | 736 | 0.179 | 0.6031 | Yes |
| 7 | ANXA3 | ANXA3 Entrez, Source | annexin A3 | 1170 | 0.131 | 0.6096 | Yes |
| 8 | ANXA5 | ANXA5 Entrez, Source | annexin A5 | 1191 | 0.129 | 0.6466 | Yes |
| 9 | ANXA6 | ANXA6 Entrez, Source | annexin A6 | 1439 | 0.111 | 0.6613 | Yes |
| 10 | HSD11B1 | HSD11B1 Entrez, Source | hydroxysteroid (11-beta) dehydrogenase 1 | 1720 | 0.096 | 0.6690 | Yes |
| 11 | PTGER2 | PTGER2 Entrez, Source | prostaglandin E receptor 2 (subtype EP2), 53kDa | 2027 | 0.084 | 0.6712 | Yes |
| 12 | TBXAS1 | TBXAS1 Entrez, Source | thromboxane A synthase 1 (platelet, cytochrome P450, family 5, subfamily A) | 2191 | 0.078 | 0.6821 | Yes |
| 13 | PTGFR | PTGFR Entrez, Source | prostaglandin F receptor (FP) | 3331 | 0.047 | 0.6105 | No |
| 14 | SCGB1A1 | SCGB1A1 Entrez, Source | secretoglobin, family 1A, member 1 (uteroglobin) | 3759 | 0.037 | 0.5896 | No |
| 15 | ANXA2 | ANXA2 Entrez, Source | annexin A2 | 3949 | 0.034 | 0.5855 | No |
| 16 | PTGER1 | PTGER1 Entrez, Source | prostaglandin E receptor 1 (subtype EP1), 42kDa | 4152 | 0.030 | 0.5793 | No |
| 17 | PTGIS | PTGIS Entrez, Source | prostaglandin I2 (prostacyclin) synthase | 4653 | 0.021 | 0.5480 | No |
| 18 | PLA2G4A | PLA2G4A Entrez, Source | phospholipase A2, group IVA (cytosolic, calcium-dependent) | 4688 | 0.020 | 0.5515 | No |
| 19 | CYP11A1 | CYP11A1 Entrez, Source | cytochrome P450, family 11, subfamily A, polypeptide 1 | 5093 | 0.014 | 0.5252 | No |
| 20 | HPGD | HPGD Entrez, Source | hydroxyprostaglandin dehydrogenase 15-(NAD) | 5728 | 0.004 | 0.4788 | No |
| 21 | PTGS1 | PTGS1 Entrez, Source | prostaglandin-endoperoxide synthase 1 (prostaglandin G/H synthase and cyclooxygenase) | 6341 | -0.005 | 0.4344 | No |
| 22 | EDNRB | EDNRB Entrez, Source | endothelin receptor type B | 6802 | -0.012 | 0.4035 | No |
| 23 | HSD11B2 | HSD11B2 Entrez, Source | hydroxysteroid (11-beta) dehydrogenase 2 | 6935 | -0.014 | 0.3978 | No |
| 24 | PTGDS | PTGDS Entrez, Source | prostaglandin D2 synthase 21kDa (brain) | 8146 | -0.033 | 0.3168 | No |
| 25 | PTGS2 | PTGS2 Entrez, Source | prostaglandin-endoperoxide synthase 2 (prostaglandin G/H synthase and cyclooxygenase) | 8937 | -0.046 | 0.2712 | No |
| 26 | EDNRA | EDNRA Entrez, Source | endothelin receptor type A | 10650 | -0.081 | 0.1669 | No |
| 27 | PRL | PRL Entrez, Source | prolactin | 11818 | -0.118 | 0.1144 | No |

