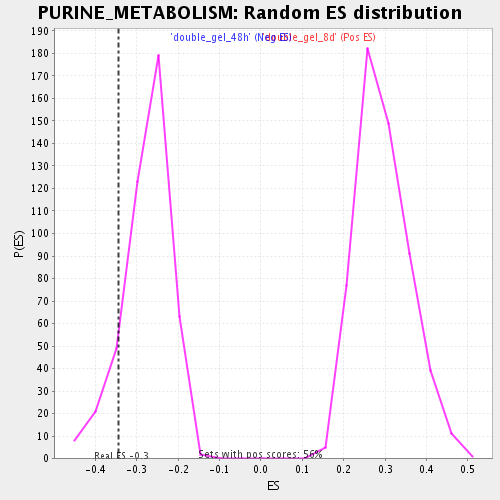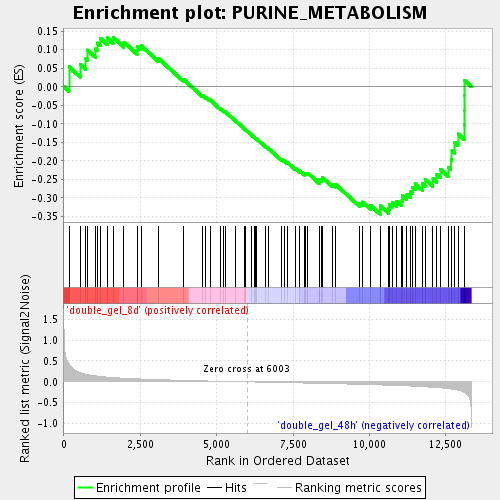
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | double_gel_and_single_gel_rma_expression_values_collapsed_to_symbols.class.cls #double_gel_8d_versus_double_gel_48h.class.cls #double_gel_8d_versus_double_gel_48h_repos |
| Phenotype | class.cls#double_gel_8d_versus_double_gel_48h_repos |
| Upregulated in class | double_gel_48h |
| GeneSet | PURINE_METABOLISM |
| Enrichment Score (ES) | -0.34354296 |
| Normalized Enrichment Score (NES) | -1.2467358 |
| Nominal p-value | 0.123595506 |
| FDR q-value | 0.52322435 |
| FWER p-Value | 1.0 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | AMPD3 | AMPD3 Entrez, Source | adenosine monophosphate deaminase (isoform E) | 167 | 0.426 | 0.0541 | No |
| 2 | GDA | GDA Entrez, Source | guanine deaminase | 537 | 0.219 | 0.0606 | No |
| 3 | PDE4C | PDE4C Entrez, Source | phosphodiesterase 4C, cAMP-specific (phosphodiesterase E1 dunce homolog, Drosophila) | 719 | 0.182 | 0.0754 | No |
| 4 | GUCY1B3 | GUCY1B3 Entrez, Source | guanylate cyclase 1, soluble, beta 3 | 766 | 0.175 | 0.0993 | No |
| 5 | POLG | POLG Entrez, Source | polymerase (DNA directed), gamma | 1028 | 0.144 | 0.1022 | No |
| 6 | GUCY2C | GUCY2C Entrez, Source | guanylate cyclase 2C (heat stable enterotoxin receptor) | 1098 | 0.136 | 0.1183 | No |
| 7 | AMPD1 | AMPD1 Entrez, Source | adenosine monophosphate deaminase 1 (isoform M) | 1199 | 0.128 | 0.1308 | No |
| 8 | ENTPD2 | ENTPD2 Entrez, Source | ectonucleoside triphosphate diphosphohydrolase 2 | 1415 | 0.113 | 0.1323 | No |
| 9 | GUK1 | GUK1 Entrez, Source | guanylate kinase 1 | 1609 | 0.102 | 0.1337 | No |
| 10 | FHIT | FHIT Entrez, Source | fragile histidine triad gene | 1965 | 0.086 | 0.1205 | No |
| 11 | ATP1B1 | ATP1B1 Entrez, Source | ATPase, Na+/K+ transporting, beta 1 polypeptide | 2395 | 0.071 | 0.0993 | No |
| 12 | ADCY6 | ADCY6 Entrez, Source | adenylate cyclase 6 | 2414 | 0.070 | 0.1089 | No |
| 13 | NUDT2 | NUDT2 Entrez, Source | nudix (nucleoside diphosphate linked moiety X)-type motif 2 | 2523 | 0.067 | 0.1113 | No |
| 14 | ADA | ADA Entrez, Source | adenosine deaminase | 3106 | 0.052 | 0.0755 | No |
| 15 | ATP5B | ATP5B Entrez, Source | ATP synthase, H+ transporting, mitochondrial F1 complex, beta polypeptide | 3912 | 0.034 | 0.0202 | No |
| 16 | AK1 | AK1 Entrez, Source | adenylate kinase 1 | 4543 | 0.022 | -0.0238 | No |
| 17 | ATP5J | ATP5J Entrez, Source | ATP synthase, H+ transporting, mitochondrial F0 complex, subunit F6 | 4641 | 0.021 | -0.0278 | No |
| 18 | ADCY5 | ADCY5 Entrez, Source | adenylate cyclase 5 | 4781 | 0.019 | -0.0353 | No |
| 19 | PDE8A | PDE8A Entrez, Source | phosphodiesterase 8A | 5133 | 0.013 | -0.0598 | No |
| 20 | GUCY1A3 | GUCY1A3 Entrez, Source | guanylate cyclase 1, soluble, alpha 3 | 5232 | 0.011 | -0.0654 | No |
| 21 | POLR2F | POLR2F Entrez, Source | polymerase (RNA) II (DNA directed) polypeptide F | 5278 | 0.010 | -0.0672 | No |
| 22 | ADK | ADK Entrez, Source | adenosine kinase | 5620 | 0.005 | -0.0920 | No |
| 23 | ATP5G3 | ATP5G3 Entrez, Source | ATP synthase, H+ transporting, mitochondrial F0 complex, subunit C3 (subunit 9) | 5923 | 0.001 | -0.1146 | No |
| 24 | POLR2G | POLR2G Entrez, Source | polymerase (RNA) II (DNA directed) polypeptide G | 5947 | 0.001 | -0.1162 | No |
| 25 | GUCY2D | GUCY2D Entrez, Source | guanylate cyclase 2D, membrane (retina-specific) | 6125 | -0.002 | -0.1292 | No |
| 26 | PDE4B | PDE4B Entrez, Source | phosphodiesterase 4B, cAMP-specific (phosphodiesterase E4 dunce homolog, Drosophila) | 6239 | -0.004 | -0.1371 | No |
| 27 | ATP5D | ATP5D Entrez, Source | ATP synthase, H+ transporting, mitochondrial F1 complex, delta subunit | 6279 | -0.004 | -0.1394 | No |
| 28 | PDE4A | PDE4A Entrez, Source | phosphodiesterase 4A, cAMP-specific (phosphodiesterase E2 dunce homolog, Drosophila) | 6308 | -0.005 | -0.1408 | No |
| 29 | GUCY1B2 | GUCY1B2 Entrez, Source | guanylate cyclase 1, soluble, beta 2 | 6602 | -0.009 | -0.1614 | No |
| 30 | ATP5I | ATP5I Entrez, Source | ATP synthase, H+ transporting, mitochondrial F0 complex, subunit E | 6710 | -0.011 | -0.1678 | No |
| 31 | ATP5F1 | ATP5F1 Entrez, Source | ATP synthase, H+ transporting, mitochondrial F0 complex, subunit B1 | 7135 | -0.017 | -0.1970 | No |
| 32 | ATP5C1 | ATP5C1 Entrez, Source | ATP synthase, H+ transporting, mitochondrial F1 complex, gamma polypeptide 1 | 7204 | -0.018 | -0.1993 | No |
| 33 | POLR2C | POLR2C Entrez, Source | polymerase (RNA) II (DNA directed) polypeptide C, 33kDa | 7301 | -0.020 | -0.2035 | No |
| 34 | ADCY2 | ADCY2 Entrez, Source | adenylate cyclase 2 (brain) | 7570 | -0.024 | -0.2199 | No |
| 35 | NT5E | NT5E Entrez, Source | 5'-nucleotidase, ecto (CD73) | 7715 | -0.026 | -0.2267 | No |
| 36 | ATP5A1 | ATP5A1 Entrez, Source | ATP synthase, H+ transporting, mitochondrial F1 complex, alpha subunit 1, cardiac muscle | 7872 | -0.028 | -0.2340 | No |
| 37 | ATP5H | ATP5H Entrez, Source | ATP synthase, H+ transporting, mitochondrial F0 complex, subunit d | 7921 | -0.029 | -0.2330 | No |
| 38 | PDE4D | PDE4D Entrez, Source | phosphodiesterase 4D, cAMP-specific (phosphodiesterase E3 dunce homolog, Drosophila) | 7982 | -0.030 | -0.2328 | No |
| 39 | ENTPD1 | ENTPD1 Entrez, Source | ectonucleoside triphosphate diphosphohydrolase 1 | 8353 | -0.037 | -0.2549 | No |
| 40 | CANT1 | CANT1 Entrez, Source | calcium activated nucleotidase 1 | 8366 | -0.037 | -0.2501 | No |
| 41 | ATP5G1 | ATP5G1 Entrez, Source | ATP synthase, H+ transporting, mitochondrial F0 complex, subunit C1 (subunit 9) | 8437 | -0.038 | -0.2494 | No |
| 42 | PDE1A | PDE1A Entrez, Source | phosphodiesterase 1A, calmodulin-dependent | 8459 | -0.038 | -0.2450 | No |
| 43 | HPRT1 | HPRT1 Entrez, Source | hypoxanthine phosphoribosyltransferase 1 (Lesch-Nyhan syndrome) | 8796 | -0.044 | -0.2635 | No |
| 44 | ADCY8 | ADCY8 Entrez, Source | adenylate cyclase 8 (brain) | 8892 | -0.045 | -0.2635 | No |
| 45 | GMPS | GMPS Entrez, Source | guanine monphosphate synthetase | 9684 | -0.060 | -0.3138 | No |
| 46 | NME2 | NME2 Entrez, Source | non-metastatic cells 2, protein (NM23B) expressed in | 9773 | -0.061 | -0.3109 | No |
| 47 | PDE9A | PDE9A Entrez, Source | phosphodiesterase 9A | 10033 | -0.067 | -0.3199 | No |
| 48 | GUCY1A2 | GUCY1A2 Entrez, Source | guanylate cyclase 1, soluble, alpha 2 | 10348 | -0.074 | -0.3320 | Yes |
| 49 | AK2 | AK2 Entrez, Source | adenylate kinase 2 | 10352 | -0.074 | -0.3207 | Yes |
| 50 | NPR2 | NPR2 Entrez, Source | natriuretic peptide receptor B/guanylate cyclase B (atrionatriuretic peptide receptor B) | 10629 | -0.081 | -0.3289 | Yes |
| 51 | ADCY3 | ADCY3 Entrez, Source | adenylate cyclase 3 | 10657 | -0.081 | -0.3182 | Yes |
| 52 | ATIC | ATIC Entrez, Source | 5-aminoimidazole-4-carboxamide ribonucleotide formyltransferase/IMP cyclohydrolase | 10742 | -0.084 | -0.3114 | Yes |
| 53 | NPR1 | NPR1 Entrez, Source | natriuretic peptide receptor A/guanylate cyclase A (atrionatriuretic peptide receptor A) | 10893 | -0.088 | -0.3089 | Yes |
| 54 | POLD1 | POLD1 Entrez, Source | polymerase (DNA directed), delta 1, catalytic subunit 125kDa | 11062 | -0.092 | -0.3072 | Yes |
| 55 | ENPP3 | ENPP3 Entrez, Source | ectonucleotide pyrophosphatase/phosphodiesterase 3 | 11082 | -0.093 | -0.2941 | Yes |
| 56 | ATP5G2 | ATP5G2 Entrez, Source | ATP synthase, H+ transporting, mitochondrial F0 complex, subunit C2 (subunit 9) | 11226 | -0.096 | -0.2898 | Yes |
| 57 | ENPP1 | ENPP1 Entrez, Source | ectonucleotide pyrophosphatase/phosphodiesterase 1 | 11335 | -0.100 | -0.2823 | Yes |
| 58 | GART | GART Entrez, Source | phosphoribosylglycinamide formyltransferase, phosphoribosylglycinamide synthetase, phosphoribosylaminoimidazole synthetase | 11406 | -0.103 | -0.2715 | Yes |
| 59 | GUCY2F | GUCY2F Entrez, Source | guanylate cyclase 2F, retinal | 11499 | -0.106 | -0.2619 | Yes |
| 60 | PAICS | PAICS Entrez, Source | phosphoribosylaminoimidazole carboxylase, phosphoribosylaminoimidazole succinocarboxamide synthetase | 11730 | -0.114 | -0.2613 | Yes |
| 61 | PRPS1 | PRPS1 Entrez, Source | phosphoribosyl pyrophosphate synthetase 1 | 11832 | -0.118 | -0.2505 | Yes |
| 62 | POLB | POLB Entrez, Source | polymerase (DNA directed), beta | 12078 | -0.131 | -0.2485 | Yes |
| 63 | POLD2 | POLD2 Entrez, Source | polymerase (DNA directed), delta 2, regulatory subunit 50kDa | 12204 | -0.137 | -0.2365 | Yes |
| 64 | NME1 | NME1 Entrez, Source | non-metastatic cells 1, protein (NM23A) expressed in | 12324 | -0.144 | -0.2229 | Yes |
| 65 | ADCY4 | ADCY4 Entrez, Source | adenylate cyclase 4 | 12581 | -0.161 | -0.2170 | Yes |
| 66 | PDE5A | PDE5A Entrez, Source | phosphodiesterase 5A, cGMP-specific | 12668 | -0.170 | -0.1969 | Yes |
| 67 | PDE7B | PDE7B Entrez, Source | phosphodiesterase 7B | 12699 | -0.173 | -0.1721 | Yes |
| 68 | POLR1B | POLR1B Entrez, Source | polymerase (RNA) I polypeptide B, 128kDa | 12791 | -0.183 | -0.1503 | Yes |
| 69 | IMPDH2 | IMPDH2 Entrez, Source | IMP (inosine monophosphate) dehydrogenase 2 | 12899 | -0.199 | -0.1271 | Yes |
| 70 | PPAT | PPAT Entrez, Source | phosphoribosyl pyrophosphate amidotransferase | 13102 | -0.253 | -0.1028 | Yes |
| 71 | PRPS2 | PRPS2 Entrez, Source | phosphoribosyl pyrophosphate synthetase 2 | 13109 | -0.256 | -0.0632 | Yes |
| 72 | PKLR | PKLR Entrez, Source | pyruvate kinase, liver and RBC | 13114 | -0.257 | -0.0233 | Yes |
| 73 | DCK | DCK Entrez, Source | deoxycytidine kinase | 13118 | -0.258 | 0.0169 | Yes |