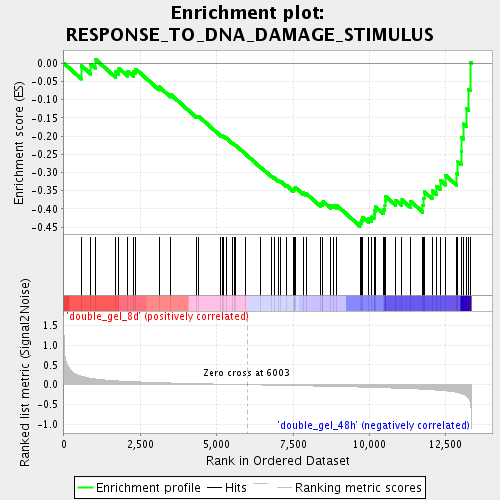
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | double_gel_and_single_gel_rma_expression_values_collapsed_to_symbols.class.cls #double_gel_8d_versus_double_gel_48h.class.cls #double_gel_8d_versus_double_gel_48h_repos |
| Phenotype | class.cls#double_gel_8d_versus_double_gel_48h_repos |
| Upregulated in class | double_gel_48h |
| GeneSet | RESPONSE_TO_DNA_DAMAGE_STIMULUS |
| Enrichment Score (ES) | -0.44733968 |
| Normalized Enrichment Score (NES) | -1.6260356 |
| Nominal p-value | 0.0070422534 |
| FDR q-value | 0.12512106 |
| FWER p-Value | 1.0 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | XRCC4 | XRCC4 Entrez, Source | X-ray repair complementing defective repair in Chinese hamster cells 4 | 575 | 0.211 | -0.0070 | No |
| 2 | BTG2 | BTG2 Entrez, Source | BTG family, member 2 | 880 | 0.159 | -0.0026 | No |
| 3 | POLG | POLG Entrez, Source | polymerase (DNA directed), gamma | 1028 | 0.144 | 0.0112 | No |
| 4 | CIB1 | CIB1 Entrez, Source | calcium and integrin binding 1 (calmyrin) | 1688 | 0.098 | -0.0216 | No |
| 5 | MAPK12 | MAPK12 Entrez, Source | mitogen-activated protein kinase 12 | 1800 | 0.093 | -0.0139 | No |
| 6 | UBE2B | UBE2B Entrez, Source | ubiquitin-conjugating enzyme E2B (RAD6 homolog) | 2086 | 0.081 | -0.0214 | No |
| 7 | RAD23A | RAD23A Entrez, Source | RAD23 homolog A (S. cerevisiae) | 2288 | 0.074 | -0.0237 | No |
| 8 | BRE | BRE Entrez, Source | brain and reproductive organ-expressed (TNFRSF1A modulator) | 2350 | 0.072 | -0.0159 | No |
| 9 | RAD17 | RAD17 Entrez, Source | RAD17 homolog (S. pombe) | 3116 | 0.051 | -0.0646 | No |
| 10 | HMGB1 | HMGB1 Entrez, Source | high-mobility group box 1 | 3490 | 0.043 | -0.0853 | No |
| 11 | CSNK1E | CSNK1E Entrez, Source | casein kinase 1, epsilon | 4346 | 0.026 | -0.1452 | No |
| 12 | MSH2 | MSH2 Entrez, Source | mutS homolog 2, colon cancer, nonpolyposis type 1 (E. coli) | 4414 | 0.025 | -0.1459 | No |
| 13 | SUMO1 | SUMO1 Entrez, Source | SMT3 suppressor of mif two 3 homolog 1 (S. cerevisiae) | 5136 | 0.013 | -0.1980 | No |
| 14 | CDKN2D | CDKN2D Entrez, Source | cyclin-dependent kinase inhibitor 2D (p19, inhibits CDK4) | 5179 | 0.012 | -0.1991 | No |
| 15 | PARP1 | PARP1 Entrez, Source | poly (ADP-ribose) polymerase family, member 1 | 5231 | 0.011 | -0.2010 | No |
| 16 | MSH3 | MSH3 Entrez, Source | mutS homolog 3 (E. coli) | 5310 | 0.010 | -0.2052 | No |
| 17 | HTATIP | HTATIP Entrez, Source | HIV-1 Tat interacting protein, 60kDa | 5528 | 0.007 | -0.2203 | No |
| 18 | NBN | NBN Entrez, Source | nibrin | 5580 | 0.006 | -0.2231 | No |
| 19 | MNAT1 | MNAT1 Entrez, Source | menage a trois homolog 1, cyclin H assembly factor (Xenopus laevis) | 5616 | 0.005 | -0.2248 | No |
| 20 | CSNK1D | CSNK1D Entrez, Source | casein kinase 1, delta | 5941 | 0.001 | -0.2490 | No |
| 21 | MAP2K6 | MAP2K6 Entrez, Source | mitogen-activated protein kinase kinase 6 | 6447 | -0.007 | -0.2859 | No |
| 22 | BRCA1 | BRCA1 Entrez, Source | breast cancer 1, early onset | 6790 | -0.012 | -0.3096 | No |
| 23 | FANCC | FANCC Entrez, Source | Fanconi anemia, complementation group C | 6887 | -0.013 | -0.3145 | No |
| 24 | VCP | VCP Entrez, Source | valosin-containing protein | 7037 | -0.016 | -0.3230 | No |
| 25 | ERCC3 | ERCC3 Entrez, Source | excision repair cross-complementing rodent repair deficiency, complementation group 3 (xeroderma pigmentosum group B complementing) | 7085 | -0.016 | -0.3237 | No |
| 26 | ATRX | ATRX Entrez, Source | alpha thalassemia/mental retardation syndrome X-linked (RAD54 homolog, S. cerevisiae) | 7295 | -0.019 | -0.3361 | No |
| 27 | IGHMBP2 | IGHMBP2 Entrez, Source | immunoglobulin mu binding protein 2 | 7514 | -0.023 | -0.3487 | No |
| 28 | UBE2N | UBE2N Entrez, Source | ubiquitin-conjugating enzyme E2N (UBC13 homolog, yeast) | 7534 | -0.023 | -0.3461 | No |
| 29 | SOD1 | SOD1 Entrez, Source | superoxide dismutase 1, soluble (amyotrophic lateral sclerosis 1 (adult)) | 7545 | -0.023 | -0.3429 | No |
| 30 | DDB1 | DDB1 Entrez, Source | damage-specific DNA binding protein 1, 127kDa | 7577 | -0.024 | -0.3411 | No |
| 31 | RECQL | RECQL Entrez, Source | RecQ protein-like (DNA helicase Q1-like) | 7826 | -0.028 | -0.3550 | No |
| 32 | SMC1A | SMC1A Entrez, Source | structural maintenance of chromosomes 1A | 7928 | -0.030 | -0.3575 | No |
| 33 | SPDYA | SPDYA Entrez, Source | speedy homolog A (Drosophila) | 8392 | -0.037 | -0.3860 | No |
| 34 | RAD50 | RAD50 Entrez, Source | RAD50 homolog (S. cerevisiae) | 8469 | -0.038 | -0.3851 | No |
| 35 | OGG1 | OGG1 Entrez, Source | 8-oxoguanine DNA glycosylase | 8478 | -0.039 | -0.3791 | No |
| 36 | CHEK2 | CHEK2 Entrez, Source | CHK2 checkpoint homolog (S. pombe) | 8735 | -0.043 | -0.3910 | No |
| 37 | CHEK1 | CHEK1 Entrez, Source | CHK1 checkpoint homolog (S. pombe) | 8812 | -0.044 | -0.3891 | No |
| 38 | CCNA2 | CCNA2 Entrez, Source | cyclin A2 | 8927 | -0.046 | -0.3898 | No |
| 39 | BRCA2 | BRCA2 Entrez, Source | breast cancer 2, early onset | 9692 | -0.060 | -0.4371 | Yes |
| 40 | RAD23B | RAD23B Entrez, Source | RAD23 homolog B (S. cerevisiae) | 9740 | -0.061 | -0.4302 | Yes |
| 41 | XRCC6 | XRCC6 Entrez, Source | X-ray repair complementing defective repair in Chinese hamster cells 6 (Ku autoantigen, 70kDa) | 9780 | -0.062 | -0.4225 | Yes |
| 42 | WRNIP1 | WRNIP1 Entrez, Source | Werner helicase interacting protein 1 | 9976 | -0.066 | -0.4258 | Yes |
| 43 | APC | APC Entrez, Source | adenomatosis polyposis coli | 10080 | -0.068 | -0.4219 | Yes |
| 44 | GADD45A | GADD45A Entrez, Source | growth arrest and DNA-damage-inducible, alpha | 10152 | -0.069 | -0.4153 | Yes |
| 45 | APEX1 | APEX1 Entrez, Source | APEX nuclease (multifunctional DNA repair enzyme) 1 | 10173 | -0.070 | -0.4048 | Yes |
| 46 | TNP1 | TNP1 Entrez, Source | transition protein 1 (during histone to protamine replacement) | 10193 | -0.070 | -0.3941 | Yes |
| 47 | MUTYH | MUTYH Entrez, Source | mutY homolog (E. coli) | 10453 | -0.076 | -0.4004 | Yes |
| 48 | MPG | MPG Entrez, Source | N-methylpurine-DNA glycosylase | 10506 | -0.078 | -0.3909 | Yes |
| 49 | UBE2A | UBE2A Entrez, Source | ubiquitin-conjugating enzyme E2A (RAD6 homolog) | 10509 | -0.078 | -0.3776 | Yes |
| 50 | PNKP | PNKP Entrez, Source | polynucleotide kinase 3'-phosphatase | 10519 | -0.078 | -0.3648 | Yes |
| 51 | LIG3 | LIG3 Entrez, Source | ligase III, DNA, ATP-dependent | 10864 | -0.087 | -0.3757 | Yes |
| 52 | POLD1 | POLD1 Entrez, Source | polymerase (DNA directed), delta 1, catalytic subunit 125kDa | 11062 | -0.092 | -0.3747 | Yes |
| 53 | TP53 | TP53 Entrez, Source | tumor protein p53 (Li-Fraumeni syndrome) | 11337 | -0.100 | -0.3781 | Yes |
| 54 | XAB2 | XAB2 Entrez, Source | XPA binding protein 2 | 11747 | -0.115 | -0.3892 | Yes |
| 55 | UNG | UNG Entrez, Source | uracil-DNA glycosylase | 11765 | -0.116 | -0.3705 | Yes |
| 56 | ATXN3 | ATXN3 Entrez, Source | ataxin 3 | 11788 | -0.116 | -0.3522 | Yes |
| 57 | GTF2H4 | GTF2H4 Entrez, Source | general transcription factor IIH, polypeptide 4, 52kDa | 12050 | -0.129 | -0.3496 | Yes |
| 58 | UBE2V2 | UBE2V2 Entrez, Source | ubiquitin-conjugating enzyme E2 variant 2 | 12194 | -0.136 | -0.3369 | Yes |
| 59 | RUVBL2 | RUVBL2 Entrez, Source | RuvB-like 2 (E. coli) | 12316 | -0.144 | -0.3213 | Yes |
| 60 | LIG1 | LIG1 Entrez, Source | ligase I, DNA, ATP-dependent | 12490 | -0.154 | -0.3078 | Yes |
| 61 | RFC3 | RFC3 Entrez, Source | replication factor C (activator 1) 3, 38kDa | 12846 | -0.192 | -0.3015 | Yes |
| 62 | TDG | TDG Entrez, Source | thymine-DNA glycosylase | 12893 | -0.199 | -0.2708 | Yes |
| 63 | FEN1 | FEN1 Entrez, Source | flap structure-specific endonuclease 1 | 13004 | -0.219 | -0.2413 | Yes |
| 64 | MCM7 | MCM7 Entrez, Source | MCM7 minichromosome maintenance deficient 7 (S. cerevisiae) | 13021 | -0.223 | -0.2041 | Yes |
| 65 | APTX | APTX Entrez, Source | aprataxin | 13079 | -0.242 | -0.1667 | Yes |
| 66 | DDIT3 | DDIT3 Entrez, Source | DNA-damage-inducible transcript 3 | 13182 | -0.289 | -0.1246 | Yes |
| 67 | MRE11A | MRE11A Entrez, Source | MRE11 meiotic recombination 11 homolog A (S. cerevisiae) | 13244 | -0.339 | -0.0708 | Yes |
| 68 | GADD45G | GADD45G Entrez, Source | growth arrest and DNA-damage-inducible, gamma | 13302 | -0.454 | 0.0030 | Yes |