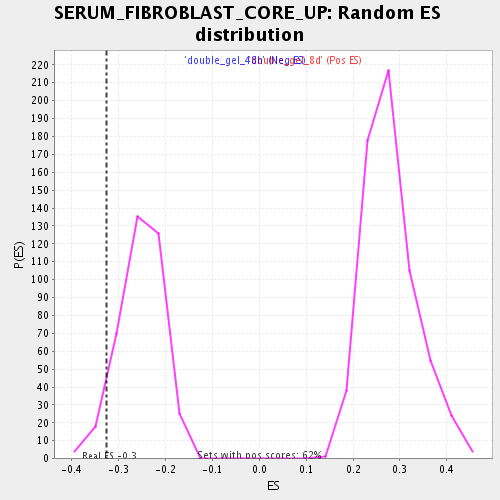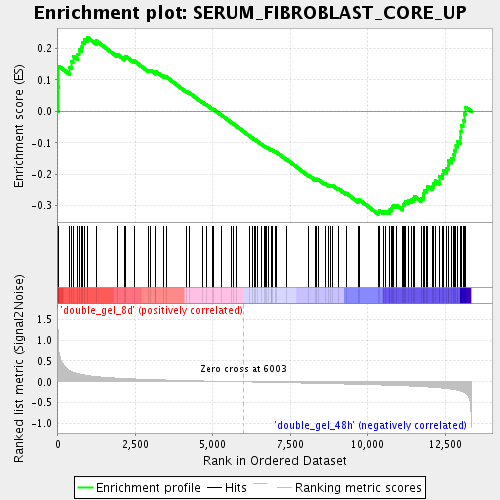
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | double_gel_and_single_gel_rma_expression_values_collapsed_to_symbols.class.cls #double_gel_8d_versus_double_gel_48h.class.cls #double_gel_8d_versus_double_gel_48h_repos |
| Phenotype | class.cls#double_gel_8d_versus_double_gel_48h_repos |
| Upregulated in class | double_gel_48h |
| GeneSet | SERUM_FIBROBLAST_CORE_UP |
| Enrichment Score (ES) | -0.32677224 |
| Normalized Enrichment Score (NES) | -1.2921456 |
| Nominal p-value | 0.05820106 |
| FDR q-value | 0.49219972 |
| FWER p-Value | 1.0 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | MAP3K8 | MAP3K8 Entrez, Source | mitogen-activated protein kinase kinase kinase 8 | 12 | 0.878 | 0.0777 | No |
| 2 | PFKP | PFKP Entrez, Source | phosphofructokinase, platelet | 29 | 0.753 | 0.1439 | No |
| 3 | TPRKB | TPRKB Entrez, Source | TP53RK binding protein | 383 | 0.267 | 0.1412 | No |
| 4 | GPLD1 | GPLD1 Entrez, Source | glycosylphosphatidylinositol specific phospholipase D1 | 445 | 0.246 | 0.1586 | No |
| 5 | TNFRSF12A | TNFRSF12A Entrez, Source | tumor necrosis factor receptor superfamily, member 12A | 489 | 0.231 | 0.1760 | No |
| 6 | CCND3 | CCND3 Entrez, Source | cyclin D3 | 643 | 0.198 | 0.1822 | No |
| 7 | SAR1B | SAR1B Entrez, Source | SAR1 gene homolog B (S. cerevisiae) | 681 | 0.188 | 0.1962 | No |
| 8 | EMP2 | EMP2 Entrez, Source | epithelial membrane protein 2 | 773 | 0.174 | 0.2049 | No |
| 9 | NUDT1 | NUDT1 Entrez, Source | nudix (nucleoside diphosphate linked moiety X)-type motif 1 | 786 | 0.172 | 0.2194 | No |
| 10 | DCBLD2 | DCBLD2 Entrez, Source | discoidin, CUB and LCCL domain containing 2 | 852 | 0.162 | 0.2290 | No |
| 11 | F3 | F3 Entrez, Source | coagulation factor III (thromboplastin, tissue factor) | 943 | 0.152 | 0.2358 | No |
| 12 | TFPI2 | TFPI2 Entrez, Source | tissue factor pathway inhibitor 2 | 1241 | 0.124 | 0.2245 | No |
| 13 | DCLRE1B | DCLRE1B Entrez, Source | DNA cross-link repair 1B (PSO2 homolog, S. cerevisiae) | 1915 | 0.088 | 0.1815 | No |
| 14 | RNF41 | RNF41 Entrez, Source | ring finger protein 41 | 2158 | 0.079 | 0.1703 | No |
| 15 | CENPJ | CENPJ Entrez, Source | centromere protein J | 2184 | 0.078 | 0.1754 | No |
| 16 | VIL2 | VIL2 Entrez, Source | villin 2 (ezrin) | 2459 | 0.069 | 0.1609 | No |
| 17 | CLCF1 | CLCF1 Entrez, Source | cardiotrophin-like cytokine factor 1 | 2926 | 0.057 | 0.1307 | No |
| 18 | PLAUR | PLAUR Entrez, Source | plasminogen activator, urokinase receptor | 2983 | 0.055 | 0.1314 | No |
| 19 | NPTN | NPTN Entrez, Source | neuroplastin | 3153 | 0.051 | 0.1232 | No |
| 20 | RNASEH2A | RNASEH2A Entrez, Source | ribonuclease H2, subunit A | 3157 | 0.051 | 0.1275 | No |
| 21 | MSN | MSN Entrez, Source | moesin | 3404 | 0.045 | 0.1129 | No |
| 22 | HMGB1 | HMGB1 Entrez, Source | high-mobility group box 1 | 3490 | 0.043 | 0.1104 | No |
| 23 | RAB3B | RAB3B Entrez, Source | RAB3B, member RAS oncogene family | 4139 | 0.030 | 0.0641 | No |
| 24 | FLNC | FLNC Entrez, Source | filamin C, gamma (actin binding protein 280) | 4230 | 0.029 | 0.0599 | No |
| 25 | PSMC3 | PSMC3 Entrez, Source | proteasome (prosome, macropain) 26S subunit, ATPase, 3 | 4668 | 0.021 | 0.0287 | No |
| 26 | COPB2 | COPB2 Entrez, Source | coatomer protein complex, subunit beta 2 (beta prime) | 4784 | 0.019 | 0.0217 | No |
| 27 | EIF4EBP1 | EIF4EBP1 Entrez, Source | eukaryotic translation initiation factor 4E binding protein 1 | 4983 | 0.015 | 0.0082 | No |
| 28 | HNRPA3 | HNRPA3 Entrez, Source | heterogeneous nuclear ribonucleoprotein A3 | 5026 | 0.015 | 0.0063 | No |
| 29 | MKKS | MKKS Entrez, Source | McKusick-Kaufman syndrome | 5274 | 0.011 | -0.0114 | No |
| 30 | MNAT1 | MNAT1 Entrez, Source | menage a trois homolog 1, cyclin H assembly factor (Xenopus laevis) | 5616 | 0.005 | -0.0367 | No |
| 31 | WSB2 | WSB2 Entrez, Source | WD repeat and SOCS box-containing 2 | 5679 | 0.005 | -0.0410 | No |
| 32 | H2AFZ | H2AFZ Entrez, Source | H2A histone family, member Z | 5766 | 0.003 | -0.0472 | No |
| 33 | MT3 | MT3 Entrez, Source | metallothionein 3 (growth inhibitory factor (neurotrophic)) | 6193 | -0.003 | -0.0791 | No |
| 34 | LYPLA2 | LYPLA2 Entrez, Source | lysophospholipase II | 6266 | -0.004 | -0.0842 | No |
| 35 | PSMD2 | PSMD2 Entrez, Source | proteasome (prosome, macropain) 26S subunit, non-ATPase, 2 | 6352 | -0.006 | -0.0901 | No |
| 36 | SSR3 | SSR3 Entrez, Source | signal sequence receptor, gamma (translocon-associated protein gamma) | 6367 | -0.006 | -0.0906 | No |
| 37 | NCLN | NCLN Entrez, Source | nicalin homolog (zebrafish) | 6432 | -0.007 | -0.0949 | No |
| 38 | PSMD12 | PSMD12 Entrez, Source | proteasome (prosome, macropain) 26S subunit, non-ATPase, 12 | 6575 | -0.009 | -0.1048 | No |
| 39 | ID3 | ID3 Entrez, Source | inhibitor of DNA binding 3, dominant negative helix-loop-helix protein | 6682 | -0.010 | -0.1119 | No |
| 40 | MCTS1 | MCTS1 Entrez, Source | malignant T cell amplified sequence 1 | 6712 | -0.011 | -0.1131 | No |
| 41 | PDLIM7 | PDLIM7 Entrez, Source | PDZ and LIM domain 7 (enigma) | 6732 | -0.011 | -0.1135 | No |
| 42 | STX3 | STX3 Entrez, Source | syntaxin 3 | 6798 | -0.012 | -0.1174 | No |
| 43 | ACTC1 | ACTC1 Entrez, Source | actin, alpha, cardiac muscle 1 | 6882 | -0.013 | -0.1224 | No |
| 44 | MAPRE1 | MAPRE1 Entrez, Source | microtubule-associated protein, RP/EB family, member 1 | 6902 | -0.014 | -0.1227 | No |
| 45 | DHFR | DHFR Entrez, Source | dihydrofolate reductase | 6938 | -0.014 | -0.1240 | No |
| 46 | PTS | PTS Entrez, Source | 6-pyruvoyltetrahydropterin synthase | 7010 | -0.015 | -0.1280 | No |
| 47 | CDCA4 | CDCA4 Entrez, Source | cell division cycle associated 4 | 7042 | -0.016 | -0.1290 | No |
| 48 | CDK2 | CDK2 Entrez, Source | cyclin-dependent kinase 2 | 7386 | -0.021 | -0.1530 | No |
| 49 | PSMA7 | PSMA7 Entrez, Source | proteasome (prosome, macropain) subunit, alpha type, 7 | 8098 | -0.032 | -0.2038 | No |
| 50 | EPHB1 | EPHB1 Entrez, Source | EPH receptor B1 | 8320 | -0.036 | -0.2173 | No |
| 51 | NLN | NLN Entrez, Source | neurolysin (metallopeptidase M3 family) | 8333 | -0.036 | -0.2150 | No |
| 52 | MRPL37 | MRPL37 Entrez, Source | mitochondrial ribosomal protein L37 | 8401 | -0.037 | -0.2167 | No |
| 53 | SLC25A5 | SLC25A5 Entrez, Source | solute carrier family 25 (mitochondrial carrier; adenine nucleotide translocator), member 5 | 8622 | -0.041 | -0.2297 | No |
| 54 | ST3GAL6 | ST3GAL6 Entrez, Source | ST3 beta-galactoside alpha-2,3-sialyltransferase 6 | 8734 | -0.043 | -0.2342 | No |
| 55 | CHEK1 | CHEK1 Entrez, Source | CHK1 checkpoint homolog (S. pombe) | 8812 | -0.044 | -0.2361 | No |
| 56 | SDC1 | SDC1 Entrez, Source | syndecan 1 | 8872 | -0.045 | -0.2365 | No |
| 57 | PCSK7 | PCSK7 Entrez, Source | proprotein convertase subtilisin/kexin type 7 | 9048 | -0.048 | -0.2454 | No |
| 58 | NUP35 | NUP35 Entrez, Source | nucleoporin 35kDa | 9299 | -0.053 | -0.2596 | No |
| 59 | BRCA2 | BRCA2 Entrez, Source | breast cancer 2, early onset | 9692 | -0.060 | -0.2839 | No |
| 60 | HNRPR | HNRPR Entrez, Source | heterogeneous nuclear ribonucleoprotein R | 9722 | -0.060 | -0.2807 | No |
| 61 | MRPL12 | MRPL12 Entrez, Source | mitochondrial ribosomal protein L12 | 10333 | -0.073 | -0.3202 | Yes |
| 62 | TXNL2 | TXNL2 Entrez, Source | thioredoxin-like 2 | 10367 | -0.074 | -0.3161 | Yes |
| 63 | SMS | SMS Entrez, Source | spermine synthase | 10501 | -0.078 | -0.3192 | Yes |
| 64 | FARSLB | FARSLB Entrez, Source | phenylalanine-tRNA synthetase-like, beta subunit | 10582 | -0.079 | -0.3181 | Yes |
| 65 | SFRS2 | SFRS2 Entrez, Source | splicing factor, arginine/serine-rich 2 | 10691 | -0.082 | -0.3189 | Yes |
| 66 | POLE3 | POLE3 Entrez, Source | polymerase (DNA directed), epsilon 3 (p17 subunit) | 10715 | -0.083 | -0.3132 | Yes |
| 67 | HAS2 | HAS2 Entrez, Source | hyaluronan synthase 2 | 10766 | -0.084 | -0.3094 | Yes |
| 68 | UMPS | UMPS Entrez, Source | uridine monophosphate synthetase (orotate phosphoribosyl transferase and orotidine-5'-decarboxylase) | 10783 | -0.085 | -0.3030 | Yes |
| 69 | PFN1 | PFN1 Entrez, Source | profilin 1 | 10821 | -0.086 | -0.2981 | Yes |
| 70 | SNRPB | SNRPB Entrez, Source | small nuclear ribonucleoprotein polypeptides B and B1 | 10937 | -0.089 | -0.2989 | Yes |
| 71 | HMGN2 | HMGN2 Entrez, Source | high-mobility group nucleosomal binding domain 2 | 11136 | -0.094 | -0.3054 | Yes |
| 72 | NDRG1 | NDRG1 Entrez, Source | N-myc downstream regulated gene 1 | 11137 | -0.094 | -0.2970 | Yes |
| 73 | LYAR | LYAR Entrez, Source | - | 11179 | -0.095 | -0.2916 | Yes |
| 74 | MTHFD1 | MTHFD1 Entrez, Source | methylenetetrahydrofolate dehydrogenase (NADP+ dependent) 1, methenyltetrahydrofolate cyclohydrolase, formyltetrahydrofolate synthetase | 11221 | -0.096 | -0.2860 | Yes |
| 75 | RPN1 | RPN1 Entrez, Source | ribophorin I | 11310 | -0.099 | -0.2838 | Yes |
| 76 | HNRPAB | HNRPAB Entrez, Source | heterogeneous nuclear ribonucleoprotein A/B | 11401 | -0.102 | -0.2815 | Yes |
| 77 | CCT5 | CCT5 Entrez, Source | chaperonin containing TCP1, subunit 5 (epsilon) | 11472 | -0.105 | -0.2774 | Yes |
| 78 | ITGA6 | ITGA6 Entrez, Source | integrin, alpha 6 | 11513 | -0.106 | -0.2709 | Yes |
| 79 | TOMM40 | TOMM40 Entrez, Source | translocase of outer mitochondrial membrane 40 homolog (yeast) | 11720 | -0.114 | -0.2762 | Yes |
| 80 | CEP78 | CEP78 Entrez, Source | centrosomal protein 78kDa | 11794 | -0.117 | -0.2713 | Yes |
| 81 | TDP1 | TDP1 Entrez, Source | tyrosyl-DNA phosphodiesterase 1 | 11807 | -0.117 | -0.2617 | Yes |
| 82 | ACTL6A | ACTL6A Entrez, Source | actin-like 6A | 11816 | -0.118 | -0.2518 | Yes |
| 83 | EIF4G1 | EIF4G1 Entrez, Source | eukaryotic translation initiation factor 4 gamma, 1 | 11911 | -0.122 | -0.2480 | Yes |
| 84 | RPP40 | RPP40 Entrez, Source | ribonuclease P 40kDa subunit | 11919 | -0.122 | -0.2376 | Yes |
| 85 | NUP93 | NUP93 Entrez, Source | nucleoporin 93kDa | 12085 | -0.131 | -0.2383 | Yes |
| 86 | FABP5 | FABP5 Entrez, Source | fatty acid binding protein 5 (psoriasis-associated) | 12121 | -0.133 | -0.2291 | Yes |
| 87 | PDIA4 | PDIA4 Entrez, Source | protein disulfide isomerase family A, member 4 | 12175 | -0.136 | -0.2210 | Yes |
| 88 | RUVBL2 | RUVBL2 Entrez, Source | RuvB-like 2 (E. coli) | 12316 | -0.144 | -0.2187 | Yes |
| 89 | WDR77 | WDR77 Entrez, Source | WD repeat domain 77 | 12317 | -0.144 | -0.2058 | Yes |
| 90 | DUT | DUT Entrez, Source | dUTP pyrophosphatase | 12414 | -0.149 | -0.1997 | Yes |
| 91 | NUP107 | NUP107 Entrez, Source | nucleoporin 107kDa | 12429 | -0.150 | -0.1874 | Yes |
| 92 | SFRS10 | SFRS10 Entrez, Source | splicing factor, arginine/serine-rich 10 (transformer 2 homolog, Drosophila) | 12549 | -0.158 | -0.1822 | Yes |
| 93 | RNF138 | RNF138 Entrez, Source | ring finger protein 138 | 12593 | -0.163 | -0.1709 | Yes |
| 94 | MET | MET Entrez, Source | met proto-oncogene (hepatocyte growth factor receptor) | 12611 | -0.164 | -0.1575 | Yes |
| 95 | NUPL1 | NUPL1 Entrez, Source | nucleoporin like 1 | 12703 | -0.173 | -0.1488 | Yes |
| 96 | ID2 | ID2 Entrez, Source | inhibitor of DNA binding 2, dominant negative helix-loop-helix protein | 12777 | -0.181 | -0.1381 | Yes |
| 97 | EBNA1BP2 | EBNA1BP2 Entrez, Source | EBNA1 binding protein 2 | 12803 | -0.186 | -0.1234 | Yes |
| 98 | RFC3 | RFC3 Entrez, Source | replication factor C (activator 1) 3, 38kDa | 12846 | -0.192 | -0.1094 | Yes |
| 99 | GGH | GGH Entrez, Source | gamma-glutamyl hydrolase (conjugase, folylpolygammaglutamyl hydrolase) | 12902 | -0.200 | -0.0957 | Yes |
| 100 | RUVBL1 | RUVBL1 Entrez, Source | RuvB-like 1 (E. coli) | 12996 | -0.217 | -0.0833 | Yes |
| 101 | GNG11 | GNG11 Entrez, Source | guanine nucleotide binding protein (G protein), gamma 11 | 13005 | -0.219 | -0.0643 | Yes |
| 102 | MCM7 | MCM7 Entrez, Source | MCM7 minichromosome maintenance deficient 7 (S. cerevisiae) | 13021 | -0.223 | -0.0455 | Yes |
| 103 | SNRPA | SNRPA Entrez, Source | small nuclear ribonucleoprotein polypeptide A | 13076 | -0.242 | -0.0279 | Yes |
| 104 | DCK | DCK Entrez, Source | deoxycytidine kinase | 13118 | -0.258 | -0.0079 | Yes |
| 105 | CCDC5 | CCDC5 Entrez, Source | coiled-coil domain containing 5 (spindle associated) | 13162 | -0.277 | 0.0136 | Yes |