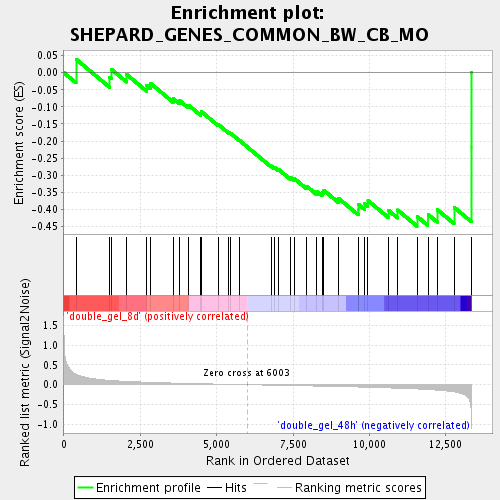
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | double_gel_and_single_gel_rma_expression_values_collapsed_to_symbols.class.cls #double_gel_8d_versus_double_gel_48h.class.cls #double_gel_8d_versus_double_gel_48h_repos |
| Phenotype | class.cls#double_gel_8d_versus_double_gel_48h_repos |
| Upregulated in class | double_gel_48h |
| GeneSet | SHEPARD_GENES_COMMON_BW_CB_MO |
| Enrichment Score (ES) | -0.44928306 |
| Normalized Enrichment Score (NES) | -1.4271518 |
| Nominal p-value | 0.054968286 |
| FDR q-value | 0.3185997 |
| FWER p-Value | 1.0 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | UCP2 | UCP2 Entrez, Source | uncoupling protein 2 (mitochondrial, proton carrier) | 398 | 0.260 | 0.0397 | No |
| 2 | RTKN | RTKN Entrez, Source | rhotekin | 1496 | 0.108 | -0.0138 | No |
| 3 | SFRP1 | SFRP1 Entrez, Source | secreted frizzled-related protein 1 | 1546 | 0.105 | 0.0107 | No |
| 4 | POU3F3 | POU3F3 Entrez, Source | POU domain, class 3, transcription factor 3 | 2053 | 0.083 | -0.0051 | No |
| 5 | TACC3 | TACC3 Entrez, Source | transforming, acidic coiled-coil containing protein 3 | 2708 | 0.062 | -0.0375 | No |
| 6 | NR2F1 | NR2F1 Entrez, Source | nuclear receptor subfamily 2, group F, member 1 | 2825 | 0.059 | -0.0304 | No |
| 7 | GOLGB1 | GOLGB1 Entrez, Source | golgi autoantigen, golgin subfamily b, macrogolgin (with transmembrane signal), 1 | 3576 | 0.041 | -0.0757 | No |
| 8 | MYH8 | MYH8 Entrez, Source | myosin, heavy chain 8, skeletal muscle, perinatal | 3786 | 0.037 | -0.0816 | No |
| 9 | ATP1A1 | ATP1A1 Entrez, Source | ATPase, Na+/K+ transporting, alpha 1 polypeptide | 4072 | 0.031 | -0.0946 | No |
| 10 | OTP | OTP Entrez, Source | orthopedia homolog (Drosophila) | 4482 | 0.024 | -0.1189 | No |
| 11 | POU3F1 | POU3F1 Entrez, Source | POU domain, class 3, transcription factor 1 | 4491 | 0.024 | -0.1132 | No |
| 12 | GRIK3 | GRIK3 Entrez, Source | glutamate receptor, ionotropic, kainate 3 | 5046 | 0.014 | -0.1510 | No |
| 13 | FOXM1 | FOXM1 Entrez, Source | forkhead box M1 | 5399 | 0.009 | -0.1751 | No |
| 14 | CTSL | CTSL Entrez, Source | cathepsin L | 5451 | 0.008 | -0.1768 | No |
| 15 | CDCA2 | CDCA2 Entrez, Source | cell division cycle associated 2 | 5754 | 0.003 | -0.1986 | No |
| 16 | CCNB1 | CCNB1 Entrez, Source | cyclin B1 | 6785 | -0.012 | -0.2729 | No |
| 17 | TM7SF2 | TM7SF2 Entrez, Source | transmembrane 7 superfamily member 2 | 6881 | -0.013 | -0.2764 | No |
| 18 | BIRC5 | BIRC5 Entrez, Source | baculoviral IAP repeat-containing 5 (survivin) | 7015 | -0.015 | -0.2824 | No |
| 19 | GPSM1 | GPSM1 Entrez, Source | G-protein signalling modulator 1 (AGS3-like, C. elegans) | 7414 | -0.021 | -0.3066 | No |
| 20 | SOX11 | SOX11 Entrez, Source | SRY (sex determining region Y)-box 11 | 7530 | -0.023 | -0.3091 | No |
| 21 | LHX1 | LHX1 Entrez, Source | LIM homeobox 1 | 7941 | -0.030 | -0.3319 | No |
| 22 | DDX17 | DDX17 Entrez, Source | DEAD (Asp-Glu-Ala-Asp) box polypeptide 17 | 8274 | -0.035 | -0.3474 | No |
| 23 | TTN | TTN Entrez, Source | titin | 8450 | -0.038 | -0.3504 | No |
| 24 | AURKB | AURKB Entrez, Source | aurora kinase B | 8506 | -0.039 | -0.3441 | No |
| 25 | NUMA1 | NUMA1 Entrez, Source | nuclear mitotic apparatus protein 1 | 8995 | -0.047 | -0.3681 | No |
| 26 | PTPRN | PTPRN Entrez, Source | protein tyrosine phosphatase, receptor type, N | 9636 | -0.058 | -0.4006 | No |
| 27 | NDE1 | NDE1 Entrez, Source | nudE nuclear distribution gene E homolog 1 (A. nidulans) | 9639 | -0.058 | -0.3851 | No |
| 28 | FGFR4 | FGFR4 Entrez, Source | fibroblast growth factor receptor 4 | 9846 | -0.063 | -0.3837 | No |
| 29 | HMGA1 | HMGA1 Entrez, Source | high mobility group AT-hook 1 | 9950 | -0.065 | -0.3739 | No |
| 30 | POU3F2 | POU3F2 Entrez, Source | POU domain, class 3, transcription factor 2 | 10612 | -0.080 | -0.4022 | No |
| 31 | MDK | MDK Entrez, Source | midkine (neurite growth-promoting factor 2) | 10921 | -0.089 | -0.4016 | No |
| 32 | BMS1L | BMS1L Entrez, Source | BMS1-like, ribosome assembly protein (yeast) | 11556 | -0.108 | -0.4204 | Yes |
| 33 | HEY1 | HEY1 Entrez, Source | hairy/enhancer-of-split related with YRPW motif 1 | 11915 | -0.122 | -0.4146 | Yes |
| 34 | CDC20 | CDC20 Entrez, Source | CDC20 cell division cycle 20 homolog (S. cerevisiae) | 12218 | -0.137 | -0.4005 | Yes |
| 35 | NOTCH2 | NOTCH2 Entrez, Source | Notch homolog 2 (Drosophila) | 12769 | -0.180 | -0.3937 | Yes |
| 36 | ASCL1 | ASCL1 Entrez, Source | achaete-scute complex-like 1 (Drosophila) | 13333 | -0.811 | -0.2192 | Yes |
| 37 | EDG1 | EDG1 Entrez, Source | endothelial differentiation, sphingolipid G-protein-coupled receptor, 1 | 13334 | -0.822 | 0.0006 | Yes |