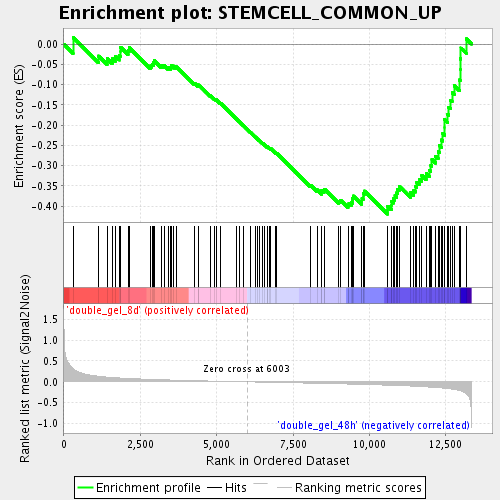
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | double_gel_and_single_gel_rma_expression_values_collapsed_to_symbols.class.cls #double_gel_8d_versus_double_gel_48h.class.cls #double_gel_8d_versus_double_gel_48h_repos |
| Phenotype | class.cls#double_gel_8d_versus_double_gel_48h_repos |
| Upregulated in class | double_gel_48h |
| GeneSet | STEMCELL_COMMON_UP |
| Enrichment Score (ES) | -0.41971645 |
| Normalized Enrichment Score (NES) | -1.6430889 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.11560792 |
| FWER p-Value | 1.0 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | SLC38A2 | SLC38A2 Entrez, Source | solute carrier family 38, member 2 | 300 | 0.310 | 0.0169 | No |
| 2 | TPP2 | TPP2 Entrez, Source | tripeptidyl peptidase II | 1139 | 0.133 | -0.0294 | No |
| 3 | PTPN2 | PTPN2 Entrez, Source | protein tyrosine phosphatase, non-receptor type 2 | 1414 | 0.113 | -0.0357 | No |
| 4 | KLHL7 | KLHL7 Entrez, Source | kelch-like 7 (Drosophila) | 1581 | 0.103 | -0.0350 | No |
| 5 | UPP1 | UPP1 Entrez, Source | uridine phosphorylase 1 | 1679 | 0.098 | -0.0298 | No |
| 6 | PANX3 | PANX3 Entrez, Source | pannexin 3 | 1807 | 0.093 | -0.0275 | No |
| 7 | PIGX | PIGX Entrez, Source | phosphatidylinositol glycan anchor biosynthesis, class X | 1844 | 0.091 | -0.0187 | No |
| 8 | PEX7 | PEX7 Entrez, Source | peroxisomal biogenesis factor 7 | 1847 | 0.091 | -0.0072 | No |
| 9 | PPP1R2 | PPP1R2 Entrez, Source | protein phosphatase 1, regulatory (inhibitor) subunit 2 | 2100 | 0.081 | -0.0159 | No |
| 10 | CTBP2 | CTBP2 Entrez, Source | C-terminal binding protein 2 | 2137 | 0.079 | -0.0085 | No |
| 11 | MPDU1 | MPDU1 Entrez, Source | mannose-P-dolichol utilization defect 1 | 2839 | 0.059 | -0.0539 | No |
| 12 | SUCLG2 | SUCLG2 Entrez, Source | succinate-CoA ligase, GDP-forming, beta subunit | 2882 | 0.058 | -0.0496 | No |
| 13 | GSTA4 | GSTA4 Entrez, Source | glutathione S-transferase A4 | 2925 | 0.057 | -0.0456 | No |
| 14 | TBC1D15 | TBC1D15 Entrez, Source | TBC1 domain family, member 15 | 2955 | 0.056 | -0.0406 | No |
| 15 | FBXO8 | FBXO8 Entrez, Source | F-box protein 8 | 3189 | 0.050 | -0.0519 | No |
| 16 | ELOVL6 | ELOVL6 Entrez, Source | ELOVL family member 6, elongation of long chain fatty acids (FEN1/Elo2, SUR4/Elo3-like, yeast) | 3278 | 0.048 | -0.0524 | No |
| 17 | SOCS2 | SOCS2 Entrez, Source | suppressor of cytokine signaling 2 | 3428 | 0.044 | -0.0580 | No |
| 18 | CTTN | CTTN Entrez, Source | cortactin | 3500 | 0.043 | -0.0579 | No |
| 19 | CDKN1A | CDKN1A Entrez, Source | cyclin-dependent kinase inhibitor 1A (p21, Cip1) | 3506 | 0.043 | -0.0528 | No |
| 20 | ALDH7A1 | ALDH7A1 Entrez, Source | aldehyde dehydrogenase 7 family, member A1 | 3592 | 0.041 | -0.0540 | No |
| 21 | KCNAB3 | KCNAB3 Entrez, Source | potassium voltage-gated channel, shaker-related subfamily, beta member 3 | 3686 | 0.039 | -0.0560 | No |
| 22 | FHL1 | FHL1 Entrez, Source | four and a half LIM domains 1 | 4269 | 0.028 | -0.0964 | No |
| 23 | YWHAB | YWHAB Entrez, Source | tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, beta polypeptide | 4394 | 0.025 | -0.1025 | No |
| 24 | MSH2 | MSH2 Entrez, Source | mutS homolog 2, colon cancer, nonpolyposis type 1 (E. coli) | 4414 | 0.025 | -0.1007 | No |
| 25 | PLS3 | PLS3 Entrez, Source | plastin 3 (T isoform) | 4795 | 0.019 | -0.1270 | No |
| 26 | ACADM | ACADM Entrez, Source | acyl-Coenzyme A dehydrogenase, C-4 to C-12 straight chain | 4941 | 0.016 | -0.1359 | No |
| 27 | EIF4EBP1 | EIF4EBP1 Entrez, Source | eukaryotic translation initiation factor 4E binding protein 1 | 4983 | 0.015 | -0.1370 | No |
| 28 | PLA2G6 | PLA2G6 Entrez, Source | phospholipase A2, group VI (cytosolic, calcium-independent) | 5121 | 0.013 | -0.1457 | No |
| 29 | ARCN1 | ARCN1 Entrez, Source | archain 1 | 5640 | 0.005 | -0.1842 | No |
| 30 | SMAD1 | SMAD1 Entrez, Source | SMAD, mothers against DPP homolog 1 (Drosophila) | 5731 | 0.004 | -0.1905 | No |
| 31 | KIF2A | KIF2A Entrez, Source | kinesin heavy chain member 2A | 5869 | 0.002 | -0.2006 | No |
| 32 | ROCK2 | ROCK2 Entrez, Source | Rho-associated, coiled-coil containing protein kinase 2 | 6100 | -0.002 | -0.2178 | No |
| 33 | STXBP3 | STXBP3 Entrez, Source | syntaxin binding protein 3 | 6260 | -0.004 | -0.2293 | No |
| 34 | RNF4 | RNF4 Entrez, Source | ring finger protein 4 | 6343 | -0.005 | -0.2348 | No |
| 35 | MTERFD1 | MTERFD1 Entrez, Source | MTERF domain containing 1 | 6399 | -0.006 | -0.2381 | No |
| 36 | TXNL1 | TXNL1 Entrez, Source | thioredoxin-like 1 | 6503 | -0.008 | -0.2449 | No |
| 37 | PSMD12 | PSMD12 Entrez, Source | proteasome (prosome, macropain) 26S subunit, non-ATPase, 12 | 6575 | -0.009 | -0.2491 | No |
| 38 | YWHAH | YWHAH Entrez, Source | tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, eta polypeptide | 6649 | -0.010 | -0.2534 | No |
| 39 | RYK | RYK Entrez, Source | RYK receptor-like tyrosine kinase | 6654 | -0.010 | -0.2525 | No |
| 40 | DNAJB6 | DNAJB6 Entrez, Source | DnaJ (Hsp40) homolog, subfamily B, member 6 | 6729 | -0.011 | -0.2566 | No |
| 41 | ITGB1 | ITGB1 Entrez, Source | integrin, beta 1 (fibronectin receptor, beta polypeptide, antigen CD29 includes MDF2, MSK12) | 6774 | -0.012 | -0.2584 | No |
| 42 | MRPL34 | MRPL34 Entrez, Source | mitochondrial ribosomal protein L34 | 6932 | -0.014 | -0.2685 | No |
| 43 | SEC23IP | SEC23IP Entrez, Source | SEC23 interacting protein | 6967 | -0.015 | -0.2692 | No |
| 44 | LAPTM4B | LAPTM4B Entrez, Source | lysosomal associated protein transmembrane 4 beta | 8069 | -0.032 | -0.3483 | No |
| 45 | MARCH7 | MARCH7 Entrez, Source | membrane-associated ring finger (C3HC4) 7 | 8284 | -0.036 | -0.3599 | No |
| 46 | SMAD2 | SMAD2 Entrez, Source | SMAD, mothers against DPP homolog 2 (Drosophila) | 8430 | -0.038 | -0.3660 | No |
| 47 | LAPTM4A | LAPTM4A Entrez, Source | lysosomal-associated protein transmembrane 4 alpha | 8436 | -0.038 | -0.3616 | No |
| 48 | TARSL1 | TARSL1 Entrez, Source | threonyl-tRNA synthetase-like 1 | 8513 | -0.039 | -0.3623 | No |
| 49 | TBRG4 | TBRG4 Entrez, Source | transforming growth factor beta regulator 4 | 8526 | -0.039 | -0.3582 | No |
| 50 | ZMAT3 | ZMAT3 Entrez, Source | zinc finger, matrin type 3 | 8994 | -0.047 | -0.3874 | No |
| 51 | UBE2D2 | UBE2D2 Entrez, Source | ubiquitin-conjugating enzyme E2D 2 (UBC4/5 homolog, yeast) | 9056 | -0.048 | -0.3859 | No |
| 52 | NUP35 | NUP35 Entrez, Source | nucleoporin 35kDa | 9299 | -0.053 | -0.3974 | No |
| 53 | MRPL17 | MRPL17 Entrez, Source | mitochondrial ribosomal protein L17 | 9327 | -0.053 | -0.3927 | No |
| 54 | XRCC5 | XRCC5 Entrez, Source | X-ray repair complementing defective repair in Chinese hamster cells 5 (double-strand-break rejoining; Ku autoantigen, 80kDa) | 9405 | -0.054 | -0.3916 | No |
| 55 | RCL1 | RCL1 Entrez, Source | RNA terminal phosphate cyclase-like 1 | 9437 | -0.055 | -0.3869 | No |
| 56 | PRPSAP1 | PRPSAP1 Entrez, Source | phosphoribosyl pyrophosphate synthetase-associated protein 1 | 9445 | -0.055 | -0.3804 | No |
| 57 | FKBP9 | FKBP9 Entrez, Source | FK506 binding protein 9, 63 kDa | 9467 | -0.055 | -0.3749 | No |
| 58 | RAD23B | RAD23B Entrez, Source | RAD23 homolog B (S. cerevisiae) | 9740 | -0.061 | -0.3877 | No |
| 59 | TXNDC9 | TXNDC9 Entrez, Source | thioredoxin domain containing 9 | 9754 | -0.061 | -0.3809 | No |
| 60 | COPS4 | COPS4 Entrez, Source | COP9 constitutive photomorphogenic homolog subunit 4 (Arabidopsis) | 9794 | -0.062 | -0.3759 | No |
| 61 | EIF4G2 | EIF4G2 Entrez, Source | eukaryotic translation initiation factor 4 gamma, 2 | 9796 | -0.062 | -0.3681 | No |
| 62 | STATIP1 | STATIP1 Entrez, Source | signal transducer and activator of transcription 3 interacting protein 1 | 9831 | -0.063 | -0.3626 | No |
| 63 | MAP4K3 | MAP4K3 Entrez, Source | mitogen-activated protein kinase kinase kinase kinase 3 | 10588 | -0.080 | -0.4096 | Yes |
| 64 | RABGGTB | RABGGTB Entrez, Source | Rab geranylgeranyltransferase, beta subunit | 10595 | -0.080 | -0.3998 | Yes |
| 65 | USP10 | USP10 Entrez, Source | ubiquitin specific peptidase 10 | 10721 | -0.083 | -0.3986 | Yes |
| 66 | ZCCHC10 | ZCCHC10 Entrez, Source | zinc finger, CCHC domain containing 10 | 10727 | -0.083 | -0.3884 | Yes |
| 67 | UMPS | UMPS Entrez, Source | uridine monophosphate synthetase (orotate phosphoribosyl transferase and orotidine-5'-decarboxylase) | 10783 | -0.085 | -0.3817 | Yes |
| 68 | CRSP3 | CRSP3 Entrez, Source | cofactor required for Sp1 transcriptional activation, subunit 3, 130kDa | 10834 | -0.086 | -0.3745 | Yes |
| 69 | STRN3 | STRN3 Entrez, Source | striatin, calmodulin binding protein 3 | 10891 | -0.088 | -0.3675 | Yes |
| 70 | IMPAD1 | IMPAD1 Entrez, Source | inositol monophosphatase domain containing 1 | 10914 | -0.088 | -0.3579 | Yes |
| 71 | GRWD1 | GRWD1 Entrez, Source | glutamate-rich WD repeat containing 1 | 10973 | -0.090 | -0.3508 | Yes |
| 72 | GARS | GARS Entrez, Source | glycyl-tRNA synthetase | 11341 | -0.100 | -0.3657 | Yes |
| 73 | LSG1 | LSG1 Entrez, Source | large subunit GTPase 1 homolog (S. cerevisiae) | 11449 | -0.104 | -0.3605 | Yes |
| 74 | ITGA6 | ITGA6 Entrez, Source | integrin, alpha 6 | 11513 | -0.106 | -0.3517 | Yes |
| 75 | ARIH1 | ARIH1 Entrez, Source | ariadne homolog, ubiquitin-conjugating enzyme E2 binding protein, 1 (Drosophila) | 11547 | -0.107 | -0.3405 | Yes |
| 76 | FKBP11 | FKBP11 Entrez, Source | FK506 binding protein 11, 19 kDa | 11643 | -0.111 | -0.3335 | Yes |
| 77 | GNB1 | GNB1 Entrez, Source | guanine nucleotide binding protein (G protein), beta polypeptide 1 | 11712 | -0.114 | -0.3241 | Yes |
| 78 | RFFL | RFFL Entrez, Source | ring finger and FYVE-like domain containing 1 | 11856 | -0.120 | -0.3197 | Yes |
| 79 | RPL22 | RPL22 Entrez, Source | ribosomal protein L22 | 11961 | -0.125 | -0.3116 | Yes |
| 80 | RPUSD4 | RPUSD4 Entrez, Source | RNA pseudouridylate synthase domain containing 4 | 12007 | -0.127 | -0.2988 | Yes |
| 81 | RPS6KB1 | RPS6KB1 Entrez, Source | ribosomal protein S6 kinase, 70kDa, polypeptide 1 | 12044 | -0.129 | -0.2851 | Yes |
| 82 | DDX1 | DDX1 Entrez, Source | DEAD (Asp-Glu-Ala-Asp) box polypeptide 1 | 12162 | -0.135 | -0.2767 | Yes |
| 83 | GCLM | GCLM Entrez, Source | glutamate-cysteine ligase, modifier subunit | 12248 | -0.139 | -0.2654 | Yes |
| 84 | MKI67IP | MKI67IP Entrez, Source | MKI67 (FHA domain) interacting nucleolar phosphoprotein | 12289 | -0.142 | -0.2503 | Yes |
| 85 | GCAT | GCAT Entrez, Source | glycine C-acetyltransferase (2-amino-3-ketobutyrate coenzyme A ligase) | 12356 | -0.146 | -0.2366 | Yes |
| 86 | PDCD2 | PDCD2 Entrez, Source | programmed cell death 2 | 12397 | -0.148 | -0.2207 | Yes |
| 87 | RASA1 | RASA1 Entrez, Source | RAS p21 protein activator (GTPase activating protein) 1 | 12457 | -0.152 | -0.2058 | Yes |
| 88 | CIRH1A | CIRH1A Entrez, Source | cirrhosis, autosomal recessive 1A (cirhin) | 12459 | -0.152 | -0.1865 | Yes |
| 89 | EPRS | EPRS Entrez, Source | glutamyl-prolyl-tRNA synthetase | 12557 | -0.159 | -0.1735 | Yes |
| 90 | RNF138 | RNF138 Entrez, Source | ring finger protein 138 | 12593 | -0.163 | -0.1554 | Yes |
| 91 | PPIC | PPIC Entrez, Source | peptidylprolyl isomerase C (cyclophilin C) | 12653 | -0.168 | -0.1383 | Yes |
| 92 | XTP3TPA | XTP3TPA Entrez, Source | - | 12705 | -0.174 | -0.1200 | Yes |
| 93 | TXNRD1 | TXNRD1 Entrez, Source | thioredoxin reductase 1 | 12775 | -0.181 | -0.1021 | Yes |
| 94 | SLC4A7 | SLC4A7 Entrez, Source | solute carrier family 4, sodium bicarbonate cotransporter, member 7 | 12948 | -0.209 | -0.0884 | Yes |
| 95 | DICER1 | DICER1 Entrez, Source | Dicer1, Dcr-1 homolog (Drosophila) | 12970 | -0.213 | -0.0628 | Yes |
| 96 | GNL2 | GNL2 Entrez, Source | guanine nucleotide binding protein-like 2 (nucleolar) | 12976 | -0.214 | -0.0359 | Yes |
| 97 | XPO1 | XPO1 Entrez, Source | exportin 1 (CRM1 homolog, yeast) | 12994 | -0.216 | -0.0096 | Yes |
| 98 | DPH5 | DPH5 Entrez, Source | DPH5 homolog (S. cerevisiae) | 13169 | -0.280 | 0.0131 | Yes |