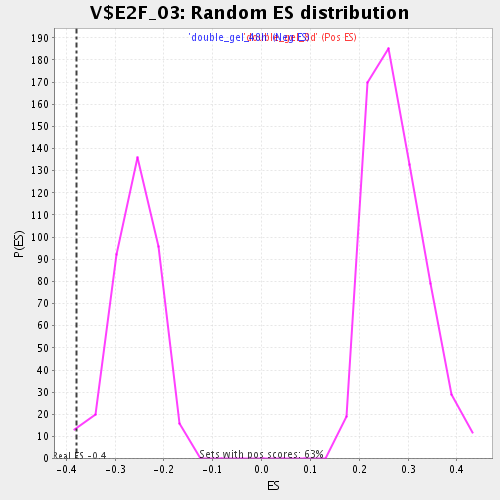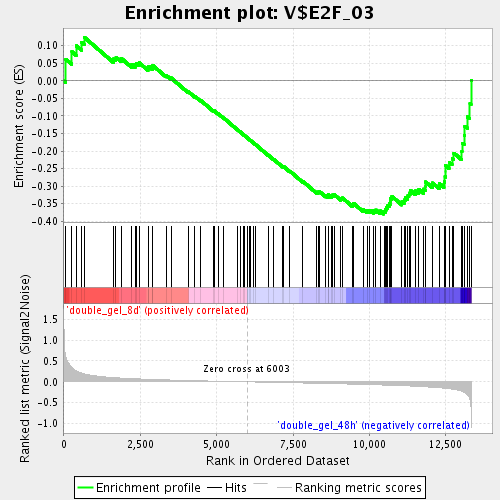
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | double_gel_and_single_gel_rma_expression_values_collapsed_to_symbols.class.cls #double_gel_8d_versus_double_gel_48h.class.cls #double_gel_8d_versus_double_gel_48h_repos |
| Phenotype | class.cls#double_gel_8d_versus_double_gel_48h_repos |
| Upregulated in class | double_gel_48h |
| GeneSet | V$E2F_03 |
| Enrichment Score (ES) | -0.37951884 |
| Normalized Enrichment Score (NES) | -1.462267 |
| Nominal p-value | 0.013404826 |
| FDR q-value | 0.27940276 |
| FWER p-Value | 1.0 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | LGALS1 | LGALS1 Entrez, Source | lectin, galactoside-binding, soluble, 1 (galectin 1) | 60 | 0.614 | 0.0618 | No |
| 2 | PAQR4 | PAQR4 Entrez, Source | progestin and adipoQ receptor family member IV | 260 | 0.335 | 0.0829 | No |
| 3 | DPYSL2 | DPYSL2 Entrez, Source | dihydropyrimidinase-like 2 | 404 | 0.258 | 0.1000 | No |
| 4 | PLAGL1 | PLAGL1 Entrez, Source | pleiomorphic adenoma gene-like 1 | 576 | 0.211 | 0.1099 | No |
| 5 | FKBP5 | FKBP5 Entrez, Source | FK506 binding protein 5 | 666 | 0.194 | 0.1242 | No |
| 6 | PCSK1 | PCSK1 Entrez, Source | proprotein convertase subtilisin/kexin type 1 | 1629 | 0.101 | 0.0625 | No |
| 7 | DAXX | DAXX Entrez, Source | death-associated protein 6 | 1703 | 0.097 | 0.0675 | No |
| 8 | SEZ6 | SEZ6 Entrez, Source | seizure related 6 homolog (mouse) | 1881 | 0.089 | 0.0638 | No |
| 9 | KLF5 | KLF5 Entrez, Source | Kruppel-like factor 5 (intestinal) | 2220 | 0.077 | 0.0466 | No |
| 10 | EGLN2 | EGLN2 Entrez, Source | egl nine homolog 2 (C. elegans) | 2331 | 0.073 | 0.0461 | No |
| 11 | PCSK4 | PCSK4 Entrez, Source | proprotein convertase subtilisin/kexin type 4 | 2377 | 0.072 | 0.0505 | No |
| 12 | ING3 | ING3 Entrez, Source | inhibitor of growth family, member 3 | 2458 | 0.069 | 0.0519 | No |
| 13 | SF4 | SF4 Entrez, Source | splicing factor 4 | 2755 | 0.061 | 0.0362 | No |
| 14 | RAB11B | RAB11B Entrez, Source | RAB11B, member RAS oncogene family | 2773 | 0.061 | 0.0415 | No |
| 15 | PPP1R9B | PPP1R9B Entrez, Source | protein phosphatase 1, regulatory subunit 9B, spinophilin | 2900 | 0.057 | 0.0382 | No |
| 16 | VAMP3 | VAMP3 Entrez, Source | vesicle-associated membrane protein 3 (cellubrevin) | 2906 | 0.057 | 0.0440 | No |
| 17 | TBX3 | TBX3 Entrez, Source | T-box 3 (ulnar mammary syndrome) | 3345 | 0.046 | 0.0159 | No |
| 18 | CDKN1A | CDKN1A Entrez, Source | cyclin-dependent kinase inhibitor 1A (p21, Cip1) | 3506 | 0.043 | 0.0085 | No |
| 19 | KCNA6 | KCNA6 Entrez, Source | potassium voltage-gated channel, shaker-related subfamily, member 6 | 4066 | 0.032 | -0.0303 | No |
| 20 | CTDSP1 | CTDSP1 Entrez, Source | CTD (carboxy-terminal domain, RNA polymerase II, polypeptide A) small phosphatase 1 | 4285 | 0.028 | -0.0438 | No |
| 21 | DIO3 | DIO3 Entrez, Source | deiodinase, iodothyronine, type III | 4469 | 0.024 | -0.0550 | No |
| 22 | KIAA0101 | KIAA0101 Entrez, Source | KIAA0101 | 4903 | 0.017 | -0.0859 | No |
| 23 | ARF3 | ARF3 Entrez, Source | ADP-ribosylation factor 3 | 4912 | 0.017 | -0.0847 | No |
| 24 | TLE3 | TLE3 Entrez, Source | transducin-like enhancer of split 3 (E(sp1) homolog, Drosophila) | 5051 | 0.014 | -0.0936 | No |
| 25 | USP52 | USP52 Entrez, Source | ubiquitin specific peptidase 52 | 5230 | 0.011 | -0.1058 | No |
| 26 | E2F1 | E2F1 Entrez, Source | E2F transcription factor 1 | 5669 | 0.005 | -0.1383 | No |
| 27 | H2AFZ | H2AFZ Entrez, Source | H2A histone family, member Z | 5766 | 0.003 | -0.1452 | No |
| 28 | DLST | DLST Entrez, Source | dihydrolipoamide S-succinyltransferase (E2 component of 2-oxo-glutarate complex) | 5892 | 0.002 | -0.1545 | No |
| 29 | GRIA4 | GRIA4 Entrez, Source | glutamate receptor, ionotrophic, AMPA 4 | 5921 | 0.001 | -0.1565 | No |
| 30 | ORC1L | ORC1L Entrez, Source | origin recognition complex, subunit 1-like (yeast) | 5994 | 0.000 | -0.1619 | No |
| 31 | CDC25A | CDC25A Entrez, Source | cell division cycle 25A | 6002 | 0.000 | -0.1624 | No |
| 32 | TOP1 | TOP1 Entrez, Source | topoisomerase (DNA) I | 6078 | -0.001 | -0.1680 | No |
| 33 | PDGFRA | PDGFRA Entrez, Source | platelet-derived growth factor receptor, alpha polypeptide | 6101 | -0.002 | -0.1694 | No |
| 34 | FBXL20 | FBXL20 Entrez, Source | F-box and leucine-rich repeat protein 20 | 6190 | -0.003 | -0.1758 | No |
| 35 | MXD3 | MXD3 Entrez, Source | MAX dimerization protein 3 | 6274 | -0.004 | -0.1816 | No |
| 36 | ID3 | ID3 Entrez, Source | inhibitor of DNA binding 3, dominant negative helix-loop-helix protein | 6682 | -0.010 | -0.2112 | No |
| 37 | CTCF | CTCF Entrez, Source | CCCTC-binding factor (zinc finger protein) | 6862 | -0.013 | -0.2233 | No |
| 38 | UXT | UXT Entrez, Source | ubiquitously-expressed transcript | 7161 | -0.018 | -0.2439 | No |
| 39 | KLHDC3 | KLHDC3 Entrez, Source | kelch domain containing 3 | 7180 | -0.018 | -0.2433 | No |
| 40 | RFC1 | RFC1 Entrez, Source | replication factor C (activator 1) 1, 145kDa | 7389 | -0.021 | -0.2568 | No |
| 41 | GLRA3 | GLRA3 Entrez, Source | glycine receptor, alpha 3 | 7823 | -0.028 | -0.2864 | No |
| 42 | DDX17 | DDX17 Entrez, Source | DEAD (Asp-Glu-Ala-Asp) box polypeptide 17 | 8274 | -0.035 | -0.3166 | No |
| 43 | EPHB1 | EPHB1 Entrez, Source | EPH receptor B1 | 8320 | -0.036 | -0.3161 | No |
| 44 | KCND2 | KCND2 Entrez, Source | potassium voltage-gated channel, Shal-related subfamily, member 2 | 8361 | -0.037 | -0.3151 | No |
| 45 | DNMT1 | DNMT1 Entrez, Source | DNA (cytosine-5-)-methyltransferase 1 | 8570 | -0.040 | -0.3265 | No |
| 46 | ANP32A | ANP32A Entrez, Source | acidic (leucine-rich) nuclear phosphoprotein 32 family, member A | 8654 | -0.041 | -0.3283 | No |
| 47 | POLA2 | POLA2 Entrez, Source | polymerase (DNA directed), alpha 2 (70kD subunit) | 8655 | -0.041 | -0.3239 | No |
| 48 | ATP6V1D | ATP6V1D Entrez, Source | ATPase, H+ transporting, lysosomal 34kDa, V1 subunit D | 8762 | -0.043 | -0.3272 | No |
| 49 | PVRL1 | PVRL1 Entrez, Source | poliovirus receptor-related 1 (herpesvirus entry mediator C; nectin) | 8795 | -0.044 | -0.3249 | No |
| 50 | NR4A2 | NR4A2 Entrez, Source | nuclear receptor subfamily 4, group A, member 2 | 8840 | -0.044 | -0.3234 | No |
| 51 | CACNA1G | CACNA1G Entrez, Source | calcium channel, voltage-dependent, alpha 1G subunit | 9066 | -0.048 | -0.3352 | No |
| 52 | TIAM1 | TIAM1 Entrez, Source | T-cell lymphoma invasion and metastasis 1 | 9112 | -0.049 | -0.3333 | No |
| 53 | ANP32E | ANP32E Entrez, Source | acidic (leucine-rich) nuclear phosphoprotein 32 family, member E | 9451 | -0.055 | -0.3528 | No |
| 54 | SLC25A11 | SLC25A11 Entrez, Source | solute carrier family 25 (mitochondrial carrier; oxoglutarate carrier), member 11 | 9476 | -0.056 | -0.3486 | No |
| 55 | EIF4G2 | EIF4G2 Entrez, Source | eukaryotic translation initiation factor 4 gamma, 2 | 9796 | -0.062 | -0.3660 | No |
| 56 | FMO4 | FMO4 Entrez, Source | flavin containing monooxygenase 4 | 9934 | -0.065 | -0.3693 | No |
| 57 | SIAHBP1 | SIAHBP1 Entrez, Source | - | 10014 | -0.067 | -0.3681 | No |
| 58 | RPL28 | RPL28 Entrez, Source | ribosomal protein L28 | 10134 | -0.069 | -0.3696 | No |
| 59 | TCP1 | TCP1 Entrez, Source | t-complex 1 | 10212 | -0.071 | -0.3678 | No |
| 60 | AK2 | AK2 Entrez, Source | adenylate kinase 2 | 10352 | -0.074 | -0.3703 | No |
| 61 | BMP7 | BMP7 Entrez, Source | bone morphogenetic protein 7 (osteogenic protein 1) | 10475 | -0.077 | -0.3712 | Yes |
| 62 | SMAD6 | SMAD6 Entrez, Source | SMAD, mothers against DPP homolog 6 (Drosophila) | 10518 | -0.078 | -0.3659 | Yes |
| 63 | MYC | MYC Entrez, Source | v-myc myelocytomatosis viral oncogene homolog (avian) | 10547 | -0.079 | -0.3595 | Yes |
| 64 | NR3C2 | NR3C2 Entrez, Source | nuclear receptor subfamily 3, group C, member 2 | 10598 | -0.080 | -0.3546 | Yes |
| 65 | STMN1 | STMN1 Entrez, Source | stathmin 1/oncoprotein 18 | 10649 | -0.081 | -0.3496 | Yes |
| 66 | RPA2 | RPA2 Entrez, Source | replication protein A2, 32kDa | 10679 | -0.082 | -0.3430 | Yes |
| 67 | SFRS2 | SFRS2 Entrez, Source | splicing factor, arginine/serine-rich 2 | 10691 | -0.082 | -0.3349 | Yes |
| 68 | HTF9C | HTF9C Entrez, Source | - | 10732 | -0.084 | -0.3289 | Yes |
| 69 | POLD1 | POLD1 Entrez, Source | polymerase (DNA directed), delta 1, catalytic subunit 125kDa | 11062 | -0.092 | -0.3438 | Yes |
| 70 | HMGN2 | HMGN2 Entrez, Source | high-mobility group nucleosomal binding domain 2 | 11136 | -0.094 | -0.3391 | Yes |
| 71 | UFD1L | UFD1L Entrez, Source | ubiquitin fusion degradation 1 like (yeast) | 11173 | -0.095 | -0.3316 | Yes |
| 72 | RET | RET Entrez, Source | ret proto-oncogene (multiple endocrine neoplasia and medullary thyroid carcinoma 1, Hirschsprung disease) | 11258 | -0.097 | -0.3274 | Yes |
| 73 | EIF5A | EIF5A Entrez, Source | eukaryotic translation initiation factor 5A | 11294 | -0.098 | -0.3194 | Yes |
| 74 | NUP153 | NUP153 Entrez, Source | nucleoporin 153kDa | 11338 | -0.100 | -0.3118 | Yes |
| 75 | RPS20 | RPS20 Entrez, Source | ribosomal protein S20 | 11511 | -0.106 | -0.3133 | Yes |
| 76 | HNRPD | HNRPD Entrez, Source | heterogeneous nuclear ribonucleoprotein D (AU-rich element RNA binding protein 1, 37kDa) | 11613 | -0.110 | -0.3091 | Yes |
| 77 | UNG | UNG Entrez, Source | uracil-DNA glycosylase | 11765 | -0.116 | -0.3080 | Yes |
| 78 | PRPS1 | PRPS1 Entrez, Source | phosphoribosyl pyrophosphate synthetase 1 | 11832 | -0.118 | -0.3002 | Yes |
| 79 | RHD | RHD Entrez, Source | Rh blood group, D antigen | 11838 | -0.119 | -0.2878 | Yes |
| 80 | EIF2S1 | EIF2S1 Entrez, Source | eukaryotic translation initiation factor 2, subunit 1 alpha, 35kDa | 12063 | -0.130 | -0.2907 | Yes |
| 81 | PRIM1 | PRIM1 Entrez, Source | primase, polypeptide 1, 49kDa | 12284 | -0.141 | -0.2920 | Yes |
| 82 | NUTF2 | NUTF2 Entrez, Source | nuclear transport factor 2 | 12441 | -0.151 | -0.2875 | Yes |
| 83 | DMD | DMD Entrez, Source | dystrophin (muscular dystrophy, Duchenne and Becker types) | 12470 | -0.153 | -0.2731 | Yes |
| 84 | PTMA | PTMA Entrez, Source | prothymosin, alpha (gene sequence 28) | 12489 | -0.154 | -0.2578 | Yes |
| 85 | LIG1 | LIG1 Entrez, Source | ligase I, DNA, ATP-dependent | 12490 | -0.154 | -0.2412 | Yes |
| 86 | PIM1 | PIM1 Entrez, Source | pim-1 oncogene | 12606 | -0.164 | -0.2322 | Yes |
| 87 | XTP3TPA | XTP3TPA Entrez, Source | - | 12705 | -0.174 | -0.2208 | Yes |
| 88 | CDCA7 | CDCA7 Entrez, Source | cell division cycle associated 7 | 12759 | -0.178 | -0.2055 | Yes |
| 89 | MCM7 | MCM7 Entrez, Source | MCM7 minichromosome maintenance deficient 7 (S. cerevisiae) | 13021 | -0.223 | -0.2011 | Yes |
| 90 | NASP | NASP Entrez, Source | nuclear autoantigenic sperm protein (histone-binding) | 13031 | -0.226 | -0.1774 | Yes |
| 91 | FUS | FUS Entrez, Source | fusion (involved in t(12;16) in malignant liposarcoma) | 13100 | -0.252 | -0.1552 | Yes |
| 92 | DCK | DCK Entrez, Source | deoxycytidine kinase | 13118 | -0.258 | -0.1286 | Yes |
| 93 | MCM6 | MCM6 Entrez, Source | MCM6 minichromosome maintenance deficient 6 (MIS5 homolog, S. pombe) (S. cerevisiae) | 13204 | -0.304 | -0.1022 | Yes |
| 94 | HNRPA1 | HNRPA1 Entrez, Source | heterogeneous nuclear ribonucleoprotein A1 | 13288 | -0.411 | -0.0641 | Yes |
| 95 | MAT2A | MAT2A Entrez, Source | methionine adenosyltransferase II, alpha | 13326 | -0.630 | 0.0012 | Yes |