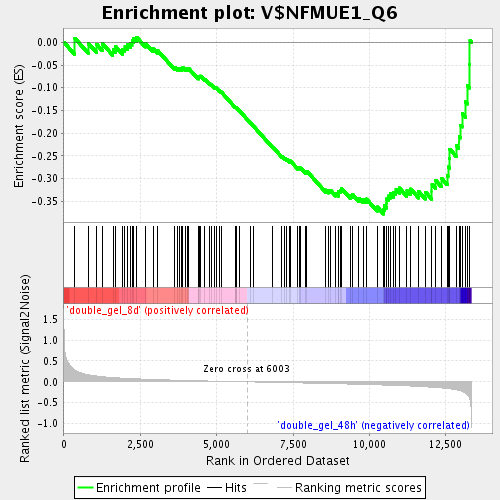
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | double_gel_and_single_gel_rma_expression_values_collapsed_to_symbols.class.cls #double_gel_8d_versus_double_gel_48h.class.cls #double_gel_8d_versus_double_gel_48h_repos |
| Phenotype | class.cls#double_gel_8d_versus_double_gel_48h_repos |
| Upregulated in class | double_gel_48h |
| GeneSet | V$NFMUE1_Q6 |
| Enrichment Score (ES) | -0.37818164 |
| Normalized Enrichment Score (NES) | -1.4786075 |
| Nominal p-value | 0.014319809 |
| FDR q-value | 0.26570916 |
| FWER p-Value | 1.0 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | NCDN | NCDN Entrez, Source | neurochondrin | 358 | 0.277 | 0.0089 | No |
| 2 | POU2F1 | POU2F1 Entrez, Source | POU domain, class 2, transcription factor 1 | 797 | 0.170 | -0.0021 | No |
| 3 | DEAF1 | DEAF1 Entrez, Source | deformed epidermal autoregulatory factor 1 (Drosophila) | 1060 | 0.140 | -0.0037 | No |
| 4 | CLK1 | CLK1 Entrez, Source | CDC-like kinase 1 | 1262 | 0.123 | -0.0030 | No |
| 5 | APBB1 | APBB1 Entrez, Source | amyloid beta (A4) precursor protein-binding, family B, member 1 (Fe65) | 1606 | 0.102 | -0.0156 | No |
| 6 | BCLAF1 | BCLAF1 Entrez, Source | BCL2-associated transcription factor 1 | 1691 | 0.098 | -0.0092 | No |
| 7 | FKRP | FKRP Entrez, Source | fukutin related protein | 1920 | 0.088 | -0.0151 | No |
| 8 | WASL | WASL Entrez, Source | Wiskott-Aldrich syndrome-like | 1998 | 0.085 | -0.0099 | No |
| 9 | VAMP2 | VAMP2 Entrez, Source | vesicle-associated membrane protein 2 (synaptobrevin 2) | 2077 | 0.082 | -0.0051 | No |
| 10 | OTX2 | OTX2 Entrez, Source | orthodenticle homolog 2 (Drosophila) | 2173 | 0.078 | -0.0022 | No |
| 11 | DZIP1 | DZIP1 Entrez, Source | DAZ interacting protein 1 | 2229 | 0.076 | 0.0036 | No |
| 12 | RAD23A | RAD23A Entrez, Source | RAD23 homolog A (S. cerevisiae) | 2288 | 0.074 | 0.0089 | No |
| 13 | UQCRH | UQCRH Entrez, Source | ubiquinol-cytochrome c reductase hinge protein | 2381 | 0.071 | 0.0112 | No |
| 14 | CCND1 | CCND1 Entrez, Source | cyclin D1 | 2668 | 0.063 | -0.0022 | No |
| 15 | CD4 | CD4 Entrez, Source | CD4 molecule | 2921 | 0.057 | -0.0139 | No |
| 16 | ZFP91 | ZFP91 Entrez, Source | zinc finger protein 91 homolog (mouse) | 3065 | 0.053 | -0.0178 | No |
| 17 | TCF4 | TCF4 Entrez, Source | transcription factor 4 | 3628 | 0.040 | -0.0550 | No |
| 18 | HOXA2 | HOXA2 Entrez, Source | homeobox A2 | 3729 | 0.038 | -0.0576 | No |
| 19 | PSMD8 | PSMD8 Entrez, Source | proteasome (prosome, macropain) 26S subunit, non-ATPase, 8 | 3796 | 0.037 | -0.0579 | No |
| 20 | ATF4 | ATF4 Entrez, Source | activating transcription factor 4 (tax-responsive enhancer element B67) | 3861 | 0.035 | -0.0581 | No |
| 21 | ZFP37 | ZFP37 Entrez, Source | zinc finger protein 37 homolog (mouse) | 3891 | 0.035 | -0.0558 | No |
| 22 | DGCR2 | DGCR2 Entrez, Source | DiGeorge syndrome critical region gene 2 | 3982 | 0.033 | -0.0583 | No |
| 23 | TIA1 | TIA1 Entrez, Source | TIA1 cytotoxic granule-associated RNA binding protein | 4046 | 0.032 | -0.0589 | No |
| 24 | ATP6V0C | ATP6V0C Entrez, Source | ATPase, H+ transporting, lysosomal 16kDa, V0 subunit c | 4083 | 0.031 | -0.0576 | No |
| 25 | MAEA | MAEA Entrez, Source | macrophage erythroblast attacher | 4393 | 0.026 | -0.0776 | No |
| 26 | PIGL | PIGL Entrez, Source | phosphatidylinositol glycan anchor biosynthesis, class L | 4412 | 0.025 | -0.0757 | No |
| 27 | SLC35A4 | SLC35A4 Entrez, Source | solute carrier family 35, member A4 | 4441 | 0.025 | -0.0746 | No |
| 28 | TSC1 | TSC1 Entrez, Source | tuberous sclerosis 1 | 4477 | 0.024 | -0.0741 | No |
| 29 | RAB1A | RAB1A Entrez, Source | RAB1A, member RAS oncogene family | 4611 | 0.021 | -0.0814 | No |
| 30 | FASTK | FASTK Entrez, Source | Fas-activated serine/threonine kinase | 4763 | 0.019 | -0.0903 | No |
| 31 | NFYA | NFYA Entrez, Source | nuclear transcription factor Y, alpha | 4833 | 0.018 | -0.0932 | No |
| 32 | PIAS3 | PIAS3 Entrez, Source | protein inhibitor of activated STAT, 3 | 4943 | 0.016 | -0.0994 | No |
| 33 | USF1 | USF1 Entrez, Source | upstream transcription factor 1 | 4980 | 0.015 | -0.1001 | No |
| 34 | MARK3 | MARK3 Entrez, Source | MAP/microtubule affinity-regulating kinase 3 | 5075 | 0.014 | -0.1054 | No |
| 35 | ARF1 | ARF1 Entrez, Source | ADP-ribosylation factor 1 | 5149 | 0.012 | -0.1093 | No |
| 36 | SNAP25 | SNAP25 Entrez, Source | synaptosomal-associated protein, 25kDa | 5609 | 0.006 | -0.1432 | No |
| 37 | ADK | ADK Entrez, Source | adenosine kinase | 5620 | 0.005 | -0.1433 | No |
| 38 | PICALM | PICALM Entrez, Source | phosphatidylinositol binding clathrin assembly protein | 5636 | 0.005 | -0.1437 | No |
| 39 | ARCN1 | ARCN1 Entrez, Source | archain 1 | 5640 | 0.005 | -0.1433 | No |
| 40 | ENSA | ENSA Entrez, Source | endosulfine alpha | 5749 | 0.003 | -0.1510 | No |
| 41 | CCNE1 | CCNE1 Entrez, Source | cyclin E1 | 6092 | -0.002 | -0.1766 | No |
| 42 | USP48 | USP48 Entrez, Source | ubiquitin specific peptidase 48 | 6200 | -0.003 | -0.1843 | No |
| 43 | RAC1 | RAC1 Entrez, Source | ras-related C3 botulinum toxin substrate 1 (rho family, small GTP binding protein Rac1) | 6834 | -0.013 | -0.2305 | No |
| 44 | ATP5F1 | ATP5F1 Entrez, Source | ATP synthase, H+ transporting, mitochondrial F0 complex, subunit B1 | 7135 | -0.017 | -0.2509 | No |
| 45 | RFX3 | RFX3 Entrez, Source | regulatory factor X, 3 (influences HLA class II expression) | 7222 | -0.019 | -0.2549 | No |
| 46 | CUGBP1 | CUGBP1 Entrez, Source | CUG triplet repeat, RNA binding protein 1 | 7294 | -0.019 | -0.2578 | No |
| 47 | APBB3 | APBB3 Entrez, Source | amyloid beta (A4) precursor protein-binding, family B, member 3 | 7383 | -0.021 | -0.2617 | No |
| 48 | NFYC | NFYC Entrez, Source | nuclear transcription factor Y, gamma | 7410 | -0.021 | -0.2610 | No |
| 49 | MID1 | MID1 Entrez, Source | midline 1 (Opitz/BBB syndrome) | 7656 | -0.025 | -0.2762 | No |
| 50 | PPP1CC | PPP1CC Entrez, Source | protein phosphatase 1, catalytic subunit, gamma isoform | 7701 | -0.026 | -0.2762 | No |
| 51 | EEF1A1 | EEF1A1 Entrez, Source | eukaryotic translation elongation factor 1 alpha 1 | 7752 | -0.027 | -0.2765 | No |
| 52 | ATP5H | ATP5H Entrez, Source | ATP synthase, H+ transporting, mitochondrial F0 complex, subunit d | 7921 | -0.029 | -0.2854 | No |
| 53 | NCOR1 | NCOR1 Entrez, Source | nuclear receptor co-repressor 1 | 7954 | -0.030 | -0.2839 | No |
| 54 | AP3D1 | AP3D1 Entrez, Source | adaptor-related protein complex 3, delta 1 subunit | 8568 | -0.040 | -0.3250 | No |
| 55 | EP300 | EP300 Entrez, Source | E1A binding protein p300 | 8660 | -0.041 | -0.3265 | No |
| 56 | NEUROD2 | NEUROD2 Entrez, Source | neurogenic differentiation 2 | 8718 | -0.043 | -0.3253 | No |
| 57 | VDAC2 | VDAC2 Entrez, Source | voltage-dependent anion channel 2 | 8895 | -0.045 | -0.3326 | No |
| 58 | RBM5 | RBM5 Entrez, Source | RNA binding motif protein 5 | 8977 | -0.047 | -0.3327 | No |
| 59 | UBE2D3 | UBE2D3 Entrez, Source | ubiquitin-conjugating enzyme E2D 3 (UBC4/5 homolog, yeast) | 8991 | -0.047 | -0.3275 | No |
| 60 | EIF3S10 | EIF3S10 Entrez, Source | eukaryotic translation initiation factor 3, subunit 10 theta, 150/170kDa | 9044 | -0.048 | -0.3252 | No |
| 61 | REST | REST Entrez, Source | RE1-silencing transcription factor | 9088 | -0.049 | -0.3222 | No |
| 62 | DNAJC7 | DNAJC7 Entrez, Source | DnaJ (Hsp40) homolog, subfamily C, member 7 | 9375 | -0.054 | -0.3368 | No |
| 63 | PRPSAP1 | PRPSAP1 Entrez, Source | phosphoribosyl pyrophosphate synthetase-associated protein 1 | 9445 | -0.055 | -0.3349 | No |
| 64 | EIF5 | EIF5 Entrez, Source | eukaryotic translation initiation factor 5 | 9653 | -0.059 | -0.3429 | No |
| 65 | EIF4G2 | EIF4G2 Entrez, Source | eukaryotic translation initiation factor 4 gamma, 2 | 9796 | -0.062 | -0.3455 | No |
| 66 | PSMB5 | PSMB5 Entrez, Source | proteasome (prosome, macropain) subunit, beta type, 5 | 9898 | -0.064 | -0.3448 | No |
| 67 | RPAP1 | RPAP1 Entrez, Source | RNA polymerase II associated protein 1 | 10254 | -0.072 | -0.3623 | No |
| 68 | FBXO11 | FBXO11 Entrez, Source | F-box protein 11 | 10465 | -0.077 | -0.3682 | Yes |
| 69 | ATF1 | ATF1 Entrez, Source | activating transcription factor 1 | 10486 | -0.077 | -0.3597 | Yes |
| 70 | CDH23 | CDH23 Entrez, Source | cadherin-like 23 | 10558 | -0.079 | -0.3548 | Yes |
| 71 | DAP3 | DAP3 Entrez, Source | death associated protein 3 | 10562 | -0.079 | -0.3447 | Yes |
| 72 | ASB2 | ASB2 Entrez, Source | ankyrin repeat and SOCS box-containing 2 | 10614 | -0.080 | -0.3382 | Yes |
| 73 | RPA2 | RPA2 Entrez, Source | replication protein A2, 32kDa | 10679 | -0.082 | -0.3324 | Yes |
| 74 | POLK | POLK Entrez, Source | polymerase (DNA directed) kappa | 10796 | -0.085 | -0.3301 | Yes |
| 75 | PABPN1 | PABPN1 Entrez, Source | poly(A) binding protein, nuclear 1 | 10867 | -0.087 | -0.3241 | Yes |
| 76 | RPL30 | RPL30 Entrez, Source | ribosomal protein L30 | 10975 | -0.090 | -0.3205 | Yes |
| 77 | ATP5G2 | ATP5G2 Entrez, Source | ATP synthase, H+ transporting, mitochondrial F0 complex, subunit C2 (subunit 9) | 11226 | -0.096 | -0.3269 | Yes |
| 78 | RPS27 | RPS27 Entrez, Source | ribosomal protein S27 (metallopanstimulin 1) | 11346 | -0.100 | -0.3228 | Yes |
| 79 | HNRPD | HNRPD Entrez, Source | heterogeneous nuclear ribonucleoprotein D (AU-rich element RNA binding protein 1, 37kDa) | 11613 | -0.110 | -0.3287 | Yes |
| 80 | PRPS1 | PRPS1 Entrez, Source | phosphoribosyl pyrophosphate synthetase 1 | 11832 | -0.118 | -0.3298 | Yes |
| 81 | RALA | RALA Entrez, Source | v-ral simian leukemia viral oncogene homolog A (ras related) | 12043 | -0.128 | -0.3290 | Yes |
| 82 | RPL35A | RPL35A Entrez, Source | ribosomal protein L35a | 12045 | -0.129 | -0.3124 | Yes |
| 83 | YY1 | YY1 Entrez, Source | YY1 transcription factor | 12166 | -0.135 | -0.3039 | Yes |
| 84 | ODF2 | ODF2 Entrez, Source | outer dense fiber of sperm tails 2 | 12355 | -0.146 | -0.2992 | Yes |
| 85 | SFRS10 | SFRS10 Entrez, Source | splicing factor, arginine/serine-rich 10 (transformer 2 homolog, Drosophila) | 12549 | -0.158 | -0.2932 | Yes |
| 86 | PTBP2 | PTBP2 Entrez, Source | polypyrimidine tract binding protein 2 | 12573 | -0.160 | -0.2742 | Yes |
| 87 | MRPL38 | MRPL38 Entrez, Source | mitochondrial ribosomal protein L38 | 12614 | -0.164 | -0.2559 | Yes |
| 88 | RBM12 | RBM12 Entrez, Source | RNA binding motif protein 12 | 12622 | -0.165 | -0.2350 | Yes |
| 89 | H1F0 | H1F0 Entrez, Source | H1 histone family, member 0 | 12841 | -0.191 | -0.2267 | Yes |
| 90 | RBM3 | RBM3 Entrez, Source | RNA binding motif (RNP1, RRM) protein 3 | 12930 | -0.204 | -0.2068 | Yes |
| 91 | GNPAT | GNPAT Entrez, Source | glyceronephosphate O-acyltransferase | 12982 | -0.215 | -0.1828 | Yes |
| 92 | NASP | NASP Entrez, Source | nuclear autoantigenic sperm protein (histone-binding) | 13031 | -0.226 | -0.1571 | Yes |
| 93 | ARNT | ARNT Entrez, Source | aryl hydrocarbon receptor nuclear translocator | 13148 | -0.269 | -0.1310 | Yes |
| 94 | SFPQ | SFPQ Entrez, Source | splicing factor proline/glutamine-rich (polypyrimidine tract binding protein associated) | 13213 | -0.310 | -0.0956 | Yes |
| 95 | CSAD | CSAD Entrez, Source | cysteine sulfinic acid decarboxylase | 13282 | -0.400 | -0.0489 | Yes |
| 96 | HNRPA1 | HNRPA1 Entrez, Source | heterogeneous nuclear ribonucleoprotein A1 | 13288 | -0.411 | 0.0041 | Yes |