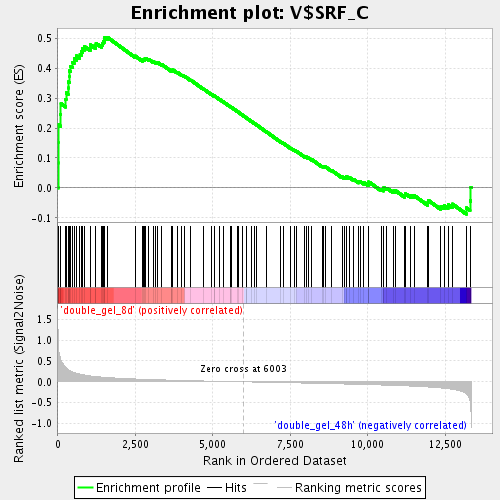
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | double_gel_and_single_gel_rma_expression_values_collapsed_to_symbols.class.cls #double_gel_8d_versus_double_gel_48h.class.cls #double_gel_8d_versus_double_gel_48h_repos |
| Phenotype | class.cls#double_gel_8d_versus_double_gel_48h_repos |
| Upregulated in class | double_gel_8d |
| GeneSet | V$SRF_C |
| Enrichment Score (ES) | 0.5039047 |
| Normalized Enrichment Score (NES) | 1.8489047 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.018633446 |
| FWER p-Value | 0.846 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | COL1A2 | COL1A2 Entrez, Source | collagen, type I, alpha 2 | 6 | 1.154 | 0.0833 | Yes |
| 2 | FOS | FOS Entrez, Source | v-fos FBJ murine osteosarcoma viral oncogene homolog | 9 | 0.936 | 0.1512 | Yes |
| 3 | SLC25A4 | SLC25A4 Entrez, Source | solute carrier family 25 (mitochondrial carrier; adenine nucleotide translocator), member 4 | 14 | 0.830 | 0.2112 | Yes |
| 4 | EGR2 | EGR2 Entrez, Source | early growth response 2 (Krox-20 homolog, Drosophila) | 87 | 0.537 | 0.2448 | Yes |
| 5 | RAB30 | RAB30 Entrez, Source | RAB30, member RAS oncogene family | 98 | 0.518 | 0.2817 | Yes |
| 6 | TAGLN | TAGLN Entrez, Source | transgelin | 250 | 0.344 | 0.2952 | Yes |
| 7 | RNF39 | RNF39 Entrez, Source | ring finger protein 39 | 266 | 0.331 | 0.3181 | Yes |
| 8 | PPP2R3A | PPP2R3A Entrez, Source | protein phosphatase 2 (formerly 2A), regulatory subunit B'', alpha | 331 | 0.289 | 0.3343 | Yes |
| 9 | RGS3 | RGS3 Entrez, Source | regulator of G-protein signalling 3 | 338 | 0.287 | 0.3547 | Yes |
| 10 | POPDC2 | POPDC2 Entrez, Source | popeye domain containing 2 | 368 | 0.273 | 0.3723 | Yes |
| 11 | FOXP1 | FOXP1 Entrez, Source | forkhead box P1 | 375 | 0.270 | 0.3915 | Yes |
| 12 | DNAJB4 | DNAJB4 Entrez, Source | DnaJ (Hsp40) homolog, subfamily B, member 4 | 410 | 0.256 | 0.4075 | Yes |
| 13 | DUSP5 | DUSP5 Entrez, Source | dual specificity phosphatase 5 | 472 | 0.237 | 0.4201 | Yes |
| 14 | LTBP1 | LTBP1 Entrez, Source | latent transforming growth factor beta binding protein 1 | 525 | 0.222 | 0.4322 | Yes |
| 15 | CYR61 | CYR61 Entrez, Source | cysteine-rich, angiogenic inducer, 61 | 600 | 0.206 | 0.4416 | Yes |
| 16 | ACTG2 | ACTG2 Entrez, Source | actin, gamma 2, smooth muscle, enteric | 711 | 0.183 | 0.4467 | Yes |
| 17 | EGR1 | EGR1 Entrez, Source | early growth response 1 | 756 | 0.176 | 0.4561 | Yes |
| 18 | SSH3 | SSH3 Entrez, Source | slingshot homolog 3 (Drosophila) | 792 | 0.170 | 0.4658 | Yes |
| 19 | JUNB | JUNB Entrez, Source | jun B proto-oncogene | 861 | 0.161 | 0.4724 | Yes |
| 20 | NR4A1 | NR4A1 Entrez, Source | nuclear receptor subfamily 4, group A, member 1 | 1043 | 0.142 | 0.4690 | Yes |
| 21 | ITGB6 | ITGB6 Entrez, Source | integrin, beta 6 | 1054 | 0.141 | 0.4785 | Yes |
| 22 | ACTA1 | ACTA1 Entrez, Source | actin, alpha 1, skeletal muscle | 1218 | 0.126 | 0.4754 | Yes |
| 23 | MYO1E | MYO1E Entrez, Source | myosin IE | 1228 | 0.126 | 0.4838 | Yes |
| 24 | PLN | PLN Entrez, Source | phospholamban | 1420 | 0.112 | 0.4775 | Yes |
| 25 | ANXA6 | ANXA6 Entrez, Source | annexin A6 | 1439 | 0.111 | 0.4842 | Yes |
| 26 | IER2 | IER2 Entrez, Source | immediate early response 2 | 1461 | 0.110 | 0.4907 | Yes |
| 27 | DUSP6 | DUSP6 Entrez, Source | dual specificity phosphatase 6 | 1504 | 0.108 | 0.4953 | Yes |
| 28 | TGFB1I1 | TGFB1I1 Entrez, Source | transforming growth factor beta 1 induced transcript 1 | 1509 | 0.108 | 0.5029 | Yes |
| 29 | MYO1B | MYO1B Entrez, Source | myosin IB | 1595 | 0.103 | 0.5039 | Yes |
| 30 | DAPK3 | DAPK3 Entrez, Source | death-associated protein kinase 3 | 2490 | 0.068 | 0.4413 | No |
| 31 | DUSP2 | DUSP2 Entrez, Source | dual specificity phosphatase 2 | 2724 | 0.062 | 0.4282 | No |
| 32 | EGR3 | EGR3 Entrez, Source | early growth response 3 | 2754 | 0.061 | 0.4305 | No |
| 33 | LCP1 | LCP1 Entrez, Source | lymphocyte cytosolic protein 1 (L-plastin) | 2802 | 0.060 | 0.4313 | No |
| 34 | NR2F1 | NR2F1 Entrez, Source | nuclear receptor subfamily 2, group F, member 1 | 2825 | 0.059 | 0.4340 | No |
| 35 | POU3F4 | POU3F4 Entrez, Source | POU domain, class 3, transcription factor 4 | 2929 | 0.057 | 0.4303 | No |
| 36 | ZFHX1B | ZFHX1B Entrez, Source | zinc finger homeobox 1b | 3081 | 0.052 | 0.4227 | No |
| 37 | PLCB3 | PLCB3 Entrez, Source | phospholipase C, beta 3 (phosphatidylinositol-specific) | 3154 | 0.051 | 0.4209 | No |
| 38 | ACTN1 | ACTN1 Entrez, Source | actinin, alpha 1 | 3213 | 0.049 | 0.4201 | No |
| 39 | MCAM | MCAM Entrez, Source | melanoma cell adhesion molecule | 3354 | 0.046 | 0.4129 | No |
| 40 | SLC2A4 | SLC2A4 Entrez, Source | solute carrier family 2 (facilitated glucose transporter), member 4 | 3677 | 0.039 | 0.3914 | No |
| 41 | ASPA | ASPA Entrez, Source | aspartoacylase (Canavan disease) | 3681 | 0.039 | 0.3940 | No |
| 42 | GPR20 | GPR20 Entrez, Source | G protein-coupled receptor 20 | 3709 | 0.039 | 0.3947 | No |
| 43 | FGFRL1 | FGFRL1 Entrez, Source | fibroblast growth factor receptor-like 1 | 3851 | 0.036 | 0.3867 | No |
| 44 | EGR4 | EGR4 Entrez, Source | early growth response 4 | 4003 | 0.033 | 0.3777 | No |
| 45 | CAP1 | CAP1 Entrez, Source | CAP, adenylate cyclase-associated protein 1 (yeast) | 4084 | 0.031 | 0.3739 | No |
| 46 | AKAP12 | AKAP12 Entrez, Source | A kinase (PRKA) anchor protein (gravin) 12 | 4283 | 0.028 | 0.3609 | No |
| 47 | KCNK3 | KCNK3 Entrez, Source | potassium channel, subfamily K, member 3 | 4699 | 0.020 | 0.3310 | No |
| 48 | GFPT2 | GFPT2 Entrez, Source | glutamine-fructose-6-phosphate transaminase 2 | 4965 | 0.016 | 0.3122 | No |
| 49 | ACTB | ACTB Entrez, Source | actin, beta | 5041 | 0.014 | 0.3075 | No |
| 50 | TLE3 | TLE3 Entrez, Source | transducin-like enhancer of split 3 (E(sp1) homolog, Drosophila) | 5051 | 0.014 | 0.3079 | No |
| 51 | USP52 | USP52 Entrez, Source | ubiquitin specific peptidase 52 | 5230 | 0.011 | 0.2953 | No |
| 52 | TCF2 | TCF2 Entrez, Source | transcription factor 2, hepatic; LF-B3; variant hepatic nuclear factor | 5337 | 0.010 | 0.2879 | No |
| 53 | MAPK14 | MAPK14 Entrez, Source | mitogen-activated protein kinase 14 | 5554 | 0.007 | 0.2721 | No |
| 54 | NR2F2 | NR2F2 Entrez, Source | nuclear receptor subfamily 2, group F, member 2 | 5565 | 0.006 | 0.2718 | No |
| 55 | RHOJ | RHOJ Entrez, Source | ras homolog gene family, member J | 5602 | 0.006 | 0.2695 | No |
| 56 | TNNC1 | TNNC1 Entrez, Source | troponin C type 1 (slow) | 5793 | 0.003 | 0.2553 | No |
| 57 | DSTN | DSTN Entrez, Source | destrin (actin depolymerizing factor) | 5815 | 0.003 | 0.2539 | No |
| 58 | CALD1 | CALD1 Entrez, Source | caldesmon 1 | 5970 | 0.001 | 0.2423 | No |
| 59 | TOP1 | TOP1 Entrez, Source | topoisomerase (DNA) I | 6078 | -0.001 | 0.2343 | No |
| 60 | ROCK2 | ROCK2 Entrez, Source | Rho-associated, coiled-coil containing protein kinase 2 | 6100 | -0.002 | 0.2329 | No |
| 61 | IL17B | IL17B Entrez, Source | interleukin 17B | 6244 | -0.004 | 0.2223 | No |
| 62 | PADI2 | PADI2 Entrez, Source | peptidyl arginine deiminase, type II | 6353 | -0.006 | 0.2146 | No |
| 63 | MBNL1 | MBNL1 Entrez, Source | muscleblind-like (Drosophila) | 6403 | -0.006 | 0.2113 | No |
| 64 | PDLIM7 | PDLIM7 Entrez, Source | PDZ and LIM domain 7 (enigma) | 6732 | -0.011 | 0.1874 | No |
| 65 | CRK | CRK Entrez, Source | v-crk sarcoma virus CT10 oncogene homolog (avian) | 7199 | -0.018 | 0.1535 | No |
| 66 | CACNA1B | CACNA1B Entrez, Source | calcium channel, voltage-dependent, L type, alpha 1B subunit | 7282 | -0.019 | 0.1487 | No |
| 67 | JPH2 | JPH2 Entrez, Source | junctophilin 2 | 7506 | -0.023 | 0.1335 | No |
| 68 | RBBP7 | RBBP7 Entrez, Source | retinoblastoma binding protein 7 | 7634 | -0.025 | 0.1257 | No |
| 69 | PPP1R12A | PPP1R12A Entrez, Source | protein phosphatase 1, regulatory (inhibitor) subunit 12A | 7696 | -0.026 | 0.1230 | No |
| 70 | ACVR1 | ACVR1 Entrez, Source | activin A receptor, type I | 7971 | -0.030 | 0.1045 | No |
| 71 | GRK6 | GRK6 Entrez, Source | G protein-coupled receptor kinase 6 | 8007 | -0.031 | 0.1041 | No |
| 72 | PDLIM4 | PDLIM4 Entrez, Source | PDZ and LIM domain 4 | 8086 | -0.032 | 0.1005 | No |
| 73 | PHF12 | PHF12 Entrez, Source | PHD finger protein 12 | 8193 | -0.034 | 0.0950 | No |
| 74 | SRD5A2 | SRD5A2 Entrez, Source | steroid-5-alpha-reductase, alpha polypeptide 2 (3-oxo-5 alpha-steroid delta 4-dehydrogenase alpha 2) | 8528 | -0.039 | 0.0726 | No |
| 75 | FOXE3 | FOXE3 Entrez, Source | forkhead box E3 | 8571 | -0.040 | 0.0723 | No |
| 76 | GGN | GGN Entrez, Source | gametogenetin | 8632 | -0.041 | 0.0707 | No |
| 77 | KCNMB1 | KCNMB1 Entrez, Source | potassium large conductance calcium-activated channel, subfamily M, beta member 1 | 8825 | -0.044 | 0.0594 | No |
| 78 | FHL2 | FHL2 Entrez, Source | four and a half LIM domains 2 | 9173 | -0.050 | 0.0369 | No |
| 79 | DLL1 | DLL1 Entrez, Source | delta-like 1 (Drosophila) | 9257 | -0.052 | 0.0344 | No |
| 80 | TITF1 | TITF1 Entrez, Source | thyroid transcription factor 1 | 9298 | -0.052 | 0.0351 | No |
| 81 | DIXDC1 | DIXDC1 Entrez, Source | DIX domain containing 1 | 9318 | -0.053 | 0.0376 | No |
| 82 | MRGPRF | MRGPRF Entrez, Source | MAS-related GPR, member F | 9403 | -0.054 | 0.0351 | No |
| 83 | MAP1A | MAP1A Entrez, Source | microtubule-associated protein 1A | 9542 | -0.057 | 0.0288 | No |
| 84 | RASD2 | RASD2 Entrez, Source | RASD family, member 2 | 9709 | -0.060 | 0.0206 | No |
| 85 | YWHAZ | YWHAZ Entrez, Source | tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, zeta polypeptide | 9753 | -0.061 | 0.0218 | No |
| 86 | ITGA7 | ITGA7 Entrez, Source | integrin, alpha 7 | 9876 | -0.064 | 0.0172 | No |
| 87 | CNN1 | CNN1 Entrez, Source | calponin 1, basic, smooth muscle | 10018 | -0.067 | 0.0114 | No |
| 88 | PRKAB2 | PRKAB2 Entrez, Source | protein kinase, AMP-activated, beta 2 non-catalytic subunit | 10029 | -0.067 | 0.0155 | No |
| 89 | MYH11 | MYH11 Entrez, Source | myosin, heavy chain 11, smooth muscle | 10036 | -0.067 | 0.0199 | No |
| 90 | AOC3 | AOC3 Entrez, Source | amine oxidase, copper containing 3 (vascular adhesion protein 1) | 10438 | -0.076 | -0.0049 | No |
| 91 | ACTR3 | ACTR3 Entrez, Source | ARP3 actin-related protein 3 homolog (yeast) | 10514 | -0.078 | -0.0048 | No |
| 92 | PNKP | PNKP Entrez, Source | polynucleotide kinase 3'-phosphatase | 10519 | -0.078 | 0.0005 | No |
| 93 | ASB2 | ASB2 Entrez, Source | ankyrin repeat and SOCS box-containing 2 | 10614 | -0.080 | -0.0007 | No |
| 94 | PFN1 | PFN1 Entrez, Source | profilin 1 | 10821 | -0.086 | -0.0101 | No |
| 95 | CFL1 | CFL1 Entrez, Source | cofilin 1 (non-muscle) | 10892 | -0.088 | -0.0090 | No |
| 96 | CKM | CKM Entrez, Source | creatine kinase, muscle | 11187 | -0.095 | -0.0242 | No |
| 97 | TPM3 | TPM3 Entrez, Source | tropomyosin 3 | 11208 | -0.096 | -0.0188 | No |
| 98 | SIPA1 | SIPA1 Entrez, Source | signal-induced proliferation-associated gene 1 | 11380 | -0.101 | -0.0243 | No |
| 99 | MUS81 | MUS81 Entrez, Source | MUS81 endonuclease homolog (S. cerevisiae) | 11503 | -0.106 | -0.0259 | No |
| 100 | FOSL1 | FOSL1 Entrez, Source | FOS-like antigen 1 | 11938 | -0.123 | -0.0497 | No |
| 101 | MATR3 | MATR3 Entrez, Source | matrin 3 | 11949 | -0.124 | -0.0414 | No |
| 102 | HOXA5 | HOXA5 Entrez, Source | homeobox A5 | 12360 | -0.146 | -0.0618 | No |
| 103 | DMD | DMD Entrez, Source | dystrophin (muscular dystrophy, Duchenne and Becker types) | 12470 | -0.153 | -0.0589 | No |
| 104 | SLC7A1 | SLC7A1 Entrez, Source | solute carrier family 7 (cationic amino acid transporter, y+ system), member 1 | 12592 | -0.163 | -0.0563 | No |
| 105 | SYNCRIP | SYNCRIP Entrez, Source | synaptotagmin binding, cytoplasmic RNA interacting protein | 12730 | -0.176 | -0.0538 | No |
| 106 | STAT5B | STAT5B Entrez, Source | signal transducer and activator of transcription 5B | 13179 | -0.287 | -0.0668 | No |
| 107 | GADD45G | GADD45G Entrez, Source | growth arrest and DNA-damage-inducible, gamma | 13302 | -0.454 | -0.0431 | No |
| 108 | CA3 | CA3 Entrez, Source | carbonic anhydrase III, muscle specific | 13327 | -0.634 | 0.0011 | No |