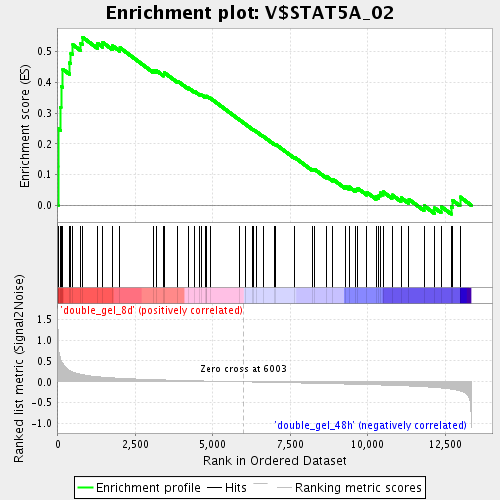
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | double_gel_and_single_gel_rma_expression_values_collapsed_to_symbols.class.cls #double_gel_8d_versus_double_gel_48h.class.cls #double_gel_8d_versus_double_gel_48h_repos |
| Phenotype | class.cls#double_gel_8d_versus_double_gel_48h_repos |
| Upregulated in class | double_gel_8d |
| GeneSet | V$STAT5A_02 |
| Enrichment Score (ES) | 0.5468269 |
| Normalized Enrichment Score (NES) | 1.8241566 |
| Nominal p-value | 0.0017667845 |
| FDR q-value | 0.022424638 |
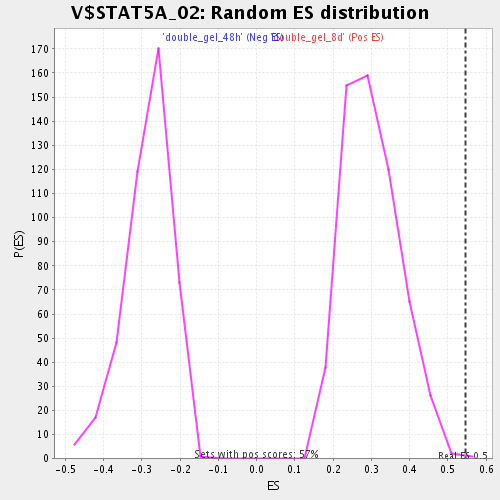
| FWER p-Value | 0.917 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | FOS | FOS Entrez, Source | v-fos FBJ murine osteosarcoma viral oncogene homolog | 9 | 0.936 | 0.1276 | Yes |
| 2 | CCND2 | CCND2 Entrez, Source | cyclin D2 | 10 | 0.885 | 0.2490 | Yes |
| 3 | PDK4 | PDK4 Entrez, Source | pyruvate dehydrogenase kinase, isozyme 4 | 84 | 0.549 | 0.3187 | Yes |
| 4 | EMP1 | EMP1 Entrez, Source | epithelial membrane protein 1 | 101 | 0.510 | 0.3873 | Yes |
| 5 | CITED4 | CITED4 Entrez, Source | Cbp/p300-interacting transactivator, with Glu/Asp-rich carboxy-terminal domain, 4 | 159 | 0.435 | 0.4426 | Yes |
| 6 | FOXP1 | FOXP1 Entrez, Source | forkhead box P1 | 375 | 0.270 | 0.4634 | Yes |
| 7 | KLF4 | KLF4 Entrez, Source | Kruppel-like factor 4 (gut) | 417 | 0.254 | 0.4952 | Yes |
| 8 | GSN | GSN Entrez, Source | gelsolin (amyloidosis, Finnish type) | 483 | 0.233 | 0.5222 | Yes |
| 9 | MAT1A | MAT1A Entrez, Source | methionine adenosyltransferase I, alpha | 734 | 0.180 | 0.5280 | Yes |
| 10 | BDNF | BDNF Entrez, Source | brain-derived neurotrophic factor | 795 | 0.170 | 0.5468 | Yes |
| 11 | PCSK2 | PCSK2 Entrez, Source | proprotein convertase subtilisin/kexin type 2 | 1278 | 0.122 | 0.5272 | No |
| 12 | SP110 | SP110 Entrez, Source | SP110 nuclear body protein | 1446 | 0.111 | 0.5299 | No |
| 13 | CACNB2 | CACNB2 Entrez, Source | calcium channel, voltage-dependent, beta 2 subunit | 1764 | 0.095 | 0.5190 | No |
| 14 | ERBB4 | ERBB4 Entrez, Source | v-erb-a erythroblastic leukemia viral oncogene homolog 4 (avian) | 2002 | 0.085 | 0.5128 | No |
| 15 | GBX2 | GBX2 Entrez, Source | gastrulation brain homeobox 2 | 3075 | 0.052 | 0.4393 | No |
| 16 | BCL2L1 | BCL2L1 Entrez, Source | BCL2-like 1 | 3166 | 0.050 | 0.4394 | No |
| 17 | NR4A3 | NR4A3 Entrez, Source | nuclear receptor subfamily 4, group A, member 3 | 3417 | 0.045 | 0.4267 | No |
| 18 | SOCS2 | SOCS2 Entrez, Source | suppressor of cytokine signaling 2 | 3428 | 0.044 | 0.4320 | No |
| 19 | ATF4 | ATF4 Entrez, Source | activating transcription factor 4 (tax-responsive enhancer element B67) | 3861 | 0.035 | 0.4044 | No |
| 20 | OBSCN | OBSCN Entrez, Source | obscurin, cytoskeletal calmodulin and titin-interacting RhoGEF | 4211 | 0.029 | 0.3821 | No |
| 21 | BTRC | BTRC Entrez, Source | beta-transducin repeat containing | 4396 | 0.025 | 0.3717 | No |
| 22 | CDC37L1 | CDC37L1 Entrez, Source | CDC37 cell division cycle 37 homolog (S. cerevisiae)-like 1 | 4571 | 0.022 | 0.3616 | No |
| 23 | ATP5J | ATP5J Entrez, Source | ATP synthase, H+ transporting, mitochondrial F0 complex, subunit F6 | 4641 | 0.021 | 0.3593 | No |
| 24 | DDX25 | DDX25 Entrez, Source | DEAD (Asp-Glu-Ala-Asp) box polypeptide 25 | 4753 | 0.019 | 0.3536 | No |
| 25 | GRID2 | GRID2 Entrez, Source | glutamate receptor, ionotropic, delta 2 | 4783 | 0.019 | 0.3540 | No |
| 26 | CREM | CREM Entrez, Source | cAMP responsive element modulator | 4794 | 0.019 | 0.3558 | No |
| 27 | ARF3 | ARF3 Entrez, Source | ADP-ribosylation factor 3 | 4912 | 0.017 | 0.3492 | No |
| 28 | GABRB1 | GABRB1 Entrez, Source | gamma-aminobutyric acid (GABA) A receptor, beta 1 | 5863 | 0.002 | 0.2780 | No |
| 29 | GPRC5C | GPRC5C Entrez, Source | G protein-coupled receptor, family C, group 5, member C | 6046 | -0.001 | 0.2644 | No |
| 30 | KCNJ8 | KCNJ8 Entrez, Source | potassium inwardly-rectifying channel, subfamily J, member 8 | 6285 | -0.004 | 0.2471 | No |
| 31 | PTPN21 | PTPN21 Entrez, Source | protein tyrosine phosphatase, non-receptor type 21 | 6317 | -0.005 | 0.2454 | No |
| 32 | MBNL1 | MBNL1 Entrez, Source | muscleblind-like (Drosophila) | 6403 | -0.006 | 0.2399 | No |
| 33 | FZD6 | FZD6 Entrez, Source | frizzled homolog 6 (Drosophila) | 6625 | -0.009 | 0.2245 | No |
| 34 | STC1 | STC1 Entrez, Source | stanniocalcin 1 | 6986 | -0.015 | 0.1995 | No |
| 35 | KITLG | KITLG Entrez, Source | KIT ligand | 7033 | -0.016 | 0.1982 | No |
| 36 | PIK3R1 | PIK3R1 Entrez, Source | phosphoinositide-3-kinase, regulatory subunit 1 (p85 alpha) | 7649 | -0.025 | 0.1553 | No |
| 37 | NTF5 | NTF5 Entrez, Source | neurotrophin 5 (neurotrophin 4/5) | 8214 | -0.034 | 0.1175 | No |
| 38 | DDX17 | DDX17 Entrez, Source | DEAD (Asp-Glu-Ala-Asp) box polypeptide 17 | 8274 | -0.035 | 0.1179 | No |
| 39 | GLRA2 | GLRA2 Entrez, Source | glycine receptor, alpha 2 | 8666 | -0.041 | 0.0942 | No |
| 40 | SDC1 | SDC1 Entrez, Source | syndecan 1 | 8872 | -0.045 | 0.0849 | No |
| 41 | VIP | VIP Entrez, Source | vasoactive intestinal peptide | 9285 | -0.052 | 0.0611 | No |
| 42 | GFAP | GFAP Entrez, Source | glial fibrillary acidic protein | 9399 | -0.054 | 0.0600 | No |
| 43 | SREBF1 | SREBF1 Entrez, Source | sterol regulatory element binding transcription factor 1 | 9603 | -0.058 | 0.0526 | No |
| 44 | RARA | RARA Entrez, Source | retinoic acid receptor, alpha | 9679 | -0.059 | 0.0551 | No |
| 45 | ARL4A | ARL4A Entrez, Source | ADP-ribosylation factor-like 4A | 9970 | -0.066 | 0.0423 | No |
| 46 | SYMPK | SYMPK Entrez, Source | symplekin | 10270 | -0.072 | 0.0296 | No |
| 47 | SRPX | SRPX Entrez, Source | sushi-repeat-containing protein, X-linked | 10360 | -0.074 | 0.0331 | No |
| 48 | IL1RN | IL1RN Entrez, Source | interleukin 1 receptor antagonist | 10398 | -0.075 | 0.0405 | No |
| 49 | LRRN3 | LRRN3 Entrez, Source | leucine rich repeat neuronal 3 | 10493 | -0.078 | 0.0441 | No |
| 50 | GJA5 | GJA5 Entrez, Source | gap junction protein, alpha 5, 40kDa (connexin 40) | 10784 | -0.085 | 0.0338 | No |
| 51 | ACOX3 | ACOX3 Entrez, Source | acyl-Coenzyme A oxidase 3, pristanoyl | 11075 | -0.092 | 0.0247 | No |
| 52 | RUNX1 | RUNX1 Entrez, Source | runt-related transcription factor 1 (acute myeloid leukemia 1; aml1 oncogene) | 11329 | -0.100 | 0.0193 | No |
| 53 | FOXA3 | FOXA3 Entrez, Source | forkhead box A3 | 11819 | -0.118 | -0.0014 | No |
| 54 | DARS | DARS Entrez, Source | aspartyl-tRNA synthetase | 12158 | -0.134 | -0.0084 | No |
| 55 | ENPP2 | ENPP2 Entrez, Source | ectonucleotide pyrophosphatase/phosphodiesterase 2 (autotaxin) | 12368 | -0.147 | -0.0040 | No |
| 56 | ZFR | ZFR Entrez, Source | zinc finger RNA binding protein | 12694 | -0.172 | -0.0049 | No |
| 57 | SYNCRIP | SYNCRIP Entrez, Source | synaptotagmin binding, cytoplasmic RNA interacting protein | 12730 | -0.176 | 0.0167 | No |
| 58 | SSBP3 | SSBP3 Entrez, Source | single stranded DNA binding protein 3 | 12979 | -0.214 | 0.0273 | No |