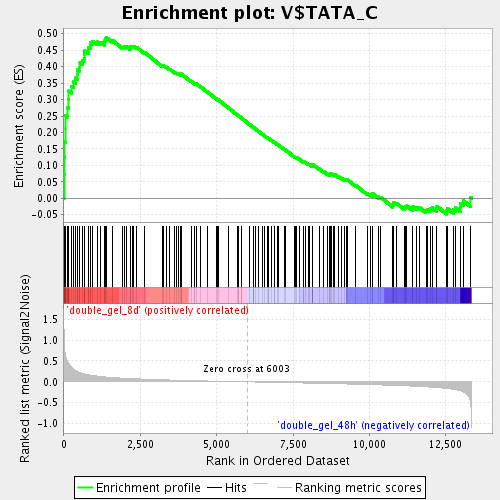
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | double_gel_and_single_gel_rma_expression_values_collapsed_to_symbols.class.cls #double_gel_8d_versus_double_gel_48h.class.cls #double_gel_8d_versus_double_gel_48h_repos |
| Phenotype | class.cls#double_gel_8d_versus_double_gel_48h_repos |
| Upregulated in class | double_gel_8d |
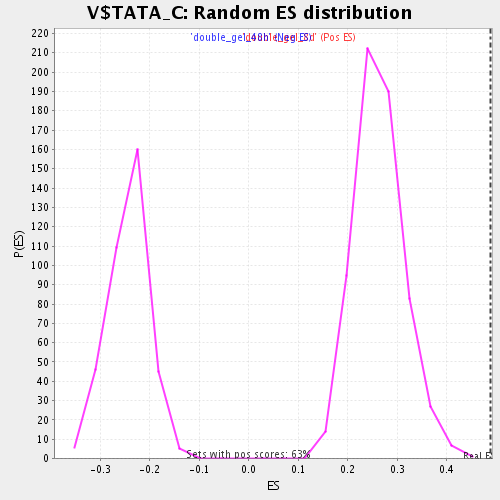
| GeneSet | V$TATA_C |
| Enrichment Score (ES) | 0.48934007 |
| Normalized Enrichment Score (NES) | 1.8582268 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.017934678 |
| FWER p-Value | 0.817 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | NPY | NPY Entrez, Source | neuropeptide Y | 5 | 1.192 | 0.0721 | Yes |
| 2 | ANXA1 | ANXA1 Entrez, Source | annexin A1 | 11 | 0.884 | 0.1255 | Yes |
| 3 | CD24 | CD24 Entrez, Source | CD24 molecule | 26 | 0.771 | 0.1714 | Yes |
| 4 | MAF | MAF Entrez, Source | v-maf musculoaponeurotic fibrosarcoma oncogene homolog (avian) | 43 | 0.675 | 0.2112 | Yes |
| 5 | LPL | LPL Entrez, Source | lipoprotein lipase | 45 | 0.671 | 0.2520 | Yes |
| 6 | GRK5 | GRK5 Entrez, Source | G protein-coupled receptor kinase 5 | 115 | 0.484 | 0.2762 | Yes |
| 7 | LOX | LOX Entrez, Source | lysyl oxidase | 154 | 0.441 | 0.3002 | Yes |
| 8 | PRKAR2B | PRKAR2B Entrez, Source | protein kinase, cAMP-dependent, regulatory, type II, beta | 160 | 0.434 | 0.3262 | Yes |
| 9 | TAGLN | TAGLN Entrez, Source | transgelin | 250 | 0.344 | 0.3404 | Yes |
| 10 | ATF3 | ATF3 Entrez, Source | activating transcription factor 3 | 310 | 0.299 | 0.3541 | Yes |
| 11 | FOXP1 | FOXP1 Entrez, Source | forkhead box P1 | 375 | 0.270 | 0.3657 | Yes |
| 12 | CYB561D2 | CYB561D2 Entrez, Source | cytochrome b-561 domain containing 2 | 443 | 0.247 | 0.3756 | Yes |
| 13 | HERPUD1 | HERPUD1 Entrez, Source | homocysteine-inducible, endoplasmic reticulum stress-inducible, ubiquitin-like domain member 1 | 448 | 0.245 | 0.3902 | Yes |
| 14 | CDC42EP2 | CDC42EP2 Entrez, Source | CDC42 effector protein (Rho GTPase binding) 2 | 522 | 0.222 | 0.3982 | Yes |
| 15 | LTBP1 | LTBP1 Entrez, Source | latent transforming growth factor beta binding protein 1 | 525 | 0.222 | 0.4116 | Yes |
| 16 | CYR61 | CYR61 Entrez, Source | cysteine-rich, angiogenic inducer, 61 | 600 | 0.206 | 0.4186 | Yes |
| 17 | TGFB2 | TGFB2 Entrez, Source | transforming growth factor, beta 2 | 659 | 0.195 | 0.4260 | Yes |
| 18 | COL1A1 | COL1A1 Entrez, Source | collagen, type I, alpha 1 | 673 | 0.190 | 0.4366 | Yes |
| 19 | CD96 | CD96 Entrez, Source | CD96 molecule | 677 | 0.189 | 0.4479 | Yes |
| 20 | BDNF | BDNF Entrez, Source | brain-derived neurotrophic factor | 795 | 0.170 | 0.4494 | Yes |
| 21 | RASL11B | RASL11B Entrez, Source | RAS-like, family 11, member B | 800 | 0.170 | 0.4594 | Yes |
| 22 | CCL7 | CCL7 Entrez, Source | chemokine (C-C motif) ligand 7 | 860 | 0.161 | 0.4648 | Yes |
| 23 | FABP4 | FABP4 Entrez, Source | fatty acid binding protein 4, adipocyte | 884 | 0.158 | 0.4726 | Yes |
| 24 | PRKAG1 | PRKAG1 Entrez, Source | protein kinase, AMP-activated, gamma 1 non-catalytic subunit | 942 | 0.152 | 0.4776 | Yes |
| 25 | TCTA | TCTA Entrez, Source | T-cell leukemia translocation altered gene | 1085 | 0.138 | 0.4752 | Yes |
| 26 | IL1B | IL1B Entrez, Source | interleukin 1, beta | 1197 | 0.128 | 0.4746 | Yes |
| 27 | EPAS1 | EPAS1 Entrez, Source | endothelial PAS domain protein 1 | 1321 | 0.118 | 0.4724 | Yes |
| 28 | PKIA | PKIA Entrez, Source | protein kinase (cAMP-dependent, catalytic) inhibitor alpha | 1340 | 0.117 | 0.4782 | Yes |
| 29 | PHEX | PHEX Entrez, Source | phosphate regulating endopeptidase homolog, X-linked (hypophosphatemia, vitamin D resistant rickets) | 1351 | 0.116 | 0.4845 | Yes |
| 30 | TWIST1 | TWIST1 Entrez, Source | twist homolog 1 (acrocephalosyndactyly 3; Saethre-Chotzen syndrome) (Drosophila) | 1380 | 0.115 | 0.4893 | Yes |
| 31 | ELMO3 | ELMO3 Entrez, Source | engulfment and cell motility 3 | 1599 | 0.103 | 0.4791 | No |
| 32 | MYH1 | MYH1 Entrez, Source | myosin, heavy chain 1, skeletal muscle, adult | 1931 | 0.087 | 0.4593 | No |
| 33 | SEMA3A | SEMA3A Entrez, Source | sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3A | 1982 | 0.086 | 0.4607 | No |
| 34 | STK39 | STK39 Entrez, Source | serine threonine kinase 39 (STE20/SPS1 homolog, yeast) | 2034 | 0.084 | 0.4620 | No |
| 35 | OTX2 | OTX2 Entrez, Source | orthodenticle homolog 2 (Drosophila) | 2173 | 0.078 | 0.4563 | No |
| 36 | MAPK10 | MAPK10 Entrez, Source | mitogen-activated protein kinase 10 | 2176 | 0.078 | 0.4609 | No |
| 37 | SDPR | SDPR Entrez, Source | serum deprivation response (phosphatidylserine binding protein) | 2231 | 0.076 | 0.4615 | No |
| 38 | SATB1 | SATB1 Entrez, Source | special AT-rich sequence binding protein 1 (binds to nuclear matrix/scaffold-associating DNA's) | 2276 | 0.075 | 0.4627 | No |
| 39 | BNIP3 | BNIP3 Entrez, Source | BCL2/adenovirus E1B 19kDa interacting protein 3 | 2391 | 0.071 | 0.4583 | No |
| 40 | PDGFRB | PDGFRB Entrez, Source | platelet-derived growth factor receptor, beta polypeptide | 2644 | 0.064 | 0.4431 | No |
| 41 | PPAP2A | PPAP2A Entrez, Source | phosphatidic acid phosphatase type 2A | 3216 | 0.049 | 0.4029 | No |
| 42 | ALDOB | ALDOB Entrez, Source | aldolase B, fructose-bisphosphate | 3253 | 0.048 | 0.4031 | No |
| 43 | TBX3 | TBX3 Entrez, Source | T-box 3 (ulnar mammary syndrome) | 3345 | 0.046 | 0.3990 | No |
| 44 | PDHA2 | PDHA2 Entrez, Source | pyruvate dehydrogenase (lipoamide) alpha 2 | 3467 | 0.044 | 0.3925 | No |
| 45 | TCF4 | TCF4 Entrez, Source | transcription factor 4 | 3628 | 0.040 | 0.3828 | No |
| 46 | TGFB3 | TGFB3 Entrez, Source | transforming growth factor, beta 3 | 3692 | 0.039 | 0.3804 | No |
| 47 | IL17F | IL17F Entrez, Source | interleukin 17F | 3733 | 0.038 | 0.3797 | No |
| 48 | RBP2 | RBP2 Entrez, Source | retinol binding protein 2, cellular | 3814 | 0.036 | 0.3758 | No |
| 49 | CDC26 | CDC26 Entrez, Source | cell division cycle 26 | 3842 | 0.036 | 0.3760 | No |
| 50 | JUND | JUND Entrez, Source | jun D proto-oncogene | 3846 | 0.036 | 0.3779 | No |
| 51 | AGPAT4 | AGPAT4 Entrez, Source | 1-acylglycerol-3-phosphate O-acyltransferase 4 (lysophosphatidic acid acyltransferase, delta) | 4160 | 0.030 | 0.3560 | No |
| 52 | AKAP12 | AKAP12 Entrez, Source | A kinase (PRKA) anchor protein (gravin) 12 | 4283 | 0.028 | 0.3485 | No |
| 53 | CTDSP1 | CTDSP1 Entrez, Source | CTD (carboxy-terminal domain, RNA polymerase II, polypeptide A) small phosphatase 1 | 4285 | 0.028 | 0.3501 | No |
| 54 | SHANK2 | SHANK2 Entrez, Source | SH3 and multiple ankyrin repeat domains 2 | 4334 | 0.027 | 0.3481 | No |
| 55 | OTP | OTP Entrez, Source | orthopedia homolog (Drosophila) | 4482 | 0.024 | 0.3384 | No |
| 56 | KCNK3 | KCNK3 Entrez, Source | potassium channel, subfamily K, member 3 | 4699 | 0.020 | 0.3232 | No |
| 57 | BARHL1 | BARHL1 Entrez, Source | BarH-like 1 (Drosophila) | 4978 | 0.016 | 0.3031 | No |
| 58 | ACTB | ACTB Entrez, Source | actin, beta | 5041 | 0.014 | 0.2993 | No |
| 59 | TLE3 | TLE3 Entrez, Source | transducin-like enhancer of split 3 (E(sp1) homolog, Drosophila) | 5051 | 0.014 | 0.2995 | No |
| 60 | NKX6-1 | NKX6-1 Entrez, Source | NK6 transcription factor related, locus 1 (Drosophila) | 5386 | 0.009 | 0.2747 | No |
| 61 | WSB2 | WSB2 Entrez, Source | WD repeat and SOCS box-containing 2 | 5679 | 0.005 | 0.2529 | No |
| 62 | NPPA | NPPA Entrez, Source | natriuretic peptide precursor A | 5715 | 0.004 | 0.2505 | No |
| 63 | DIO2 | DIO2 Entrez, Source | deiodinase, iodothyronine, type II | 5720 | 0.004 | 0.2504 | No |
| 64 | TSHB | TSHB Entrez, Source | thyroid stimulating hormone, beta | 5796 | 0.003 | 0.2449 | No |
| 65 | GNAZ | GNAZ Entrez, Source | guanine nucleotide binding protein (G protein), alpha z polypeptide | 5810 | 0.003 | 0.2441 | No |
| 66 | H3F3B | H3F3B Entrez, Source | H3 histone, family 3B (H3.3B) | 6064 | -0.001 | 0.2250 | No |
| 67 | MT3 | MT3 Entrez, Source | metallothionein 3 (growth inhibitory factor (neurotrophic)) | 6193 | -0.003 | 0.2155 | No |
| 68 | KCNA3 | KCNA3 Entrez, Source | potassium voltage-gated channel, shaker-related subfamily, member 3 | 6204 | -0.003 | 0.2149 | No |
| 69 | KCNK12 | KCNK12 Entrez, Source | potassium channel, subfamily K, member 12 | 6207 | -0.003 | 0.2149 | No |
| 70 | KCNJ8 | KCNJ8 Entrez, Source | potassium inwardly-rectifying channel, subfamily J, member 8 | 6285 | -0.004 | 0.2094 | No |
| 71 | SFRS12 | SFRS12 Entrez, Source | splicing factor, arginine/serine-rich 12 | 6368 | -0.006 | 0.2035 | No |
| 72 | CRHBP | CRHBP Entrez, Source | corticotropin releasing hormone binding protein | 6512 | -0.008 | 0.1932 | No |
| 73 | LAPTM5 | LAPTM5 Entrez, Source | lysosomal associated multispanning membrane protein 5 | 6572 | -0.009 | 0.1892 | No |
| 74 | MMP10 | MMP10 Entrez, Source | matrix metallopeptidase 10 (stromelysin 2) | 6646 | -0.010 | 0.1843 | No |
| 75 | HAMP | HAMP Entrez, Source | hepcidin antimicrobial peptide | 6695 | -0.011 | 0.1813 | No |
| 76 | MYH3 | MYH3 Entrez, Source | myosin, heavy chain 3, skeletal muscle, embryonic | 6704 | -0.011 | 0.1813 | No |
| 77 | DUOX2 | DUOX2 Entrez, Source | dual oxidase 2 | 6791 | -0.012 | 0.1755 | No |
| 78 | CRABP2 | CRABP2 Entrez, Source | cellular retinoic acid binding protein 2 | 6879 | -0.013 | 0.1698 | No |
| 79 | MAPRE1 | MAPRE1 Entrez, Source | microtubule-associated protein, RP/EB family, member 1 | 6902 | -0.014 | 0.1689 | No |
| 80 | STC1 | STC1 Entrez, Source | stanniocalcin 1 | 6986 | -0.015 | 0.1635 | No |
| 81 | KITLG | KITLG Entrez, Source | KIT ligand | 7033 | -0.016 | 0.1610 | No |
| 82 | EIF4A2 | EIF4A2 Entrez, Source | eukaryotic translation initiation factor 4A, isoform 2 | 7219 | -0.018 | 0.1481 | No |
| 83 | FSHB | FSHB Entrez, Source | follicle stimulating hormone, beta polypeptide | 7237 | -0.019 | 0.1480 | No |
| 84 | RHOH | RHOH Entrez, Source | ras homolog gene family, member H | 7559 | -0.024 | 0.1251 | No |
| 85 | H2AFX | H2AFX Entrez, Source | H2A histone family, member X | 7594 | -0.024 | 0.1240 | No |
| 86 | RHOA | RHOA Entrez, Source | ras homolog gene family, member A | 7625 | -0.025 | 0.1232 | No |
| 87 | PPP1R12A | PPP1R12A Entrez, Source | protein phosphatase 1, regulatory (inhibitor) subunit 12A | 7696 | -0.026 | 0.1195 | No |
| 88 | EEF2 | EEF2 Entrez, Source | eukaryotic translation elongation factor 2 | 7846 | -0.028 | 0.1099 | No |
| 89 | MYH2 | MYH2 Entrez, Source | myosin, heavy chain 2, skeletal muscle, adult | 7853 | -0.028 | 0.1111 | No |
| 90 | PPARGC1B | PPARGC1B Entrez, Source | peroxisome proliferative activated receptor, gamma, coactivator 1, beta | 7905 | -0.029 | 0.1091 | No |
| 91 | GPR50 | GPR50 Entrez, Source | G protein-coupled receptor 50 | 8008 | -0.031 | 0.1032 | No |
| 92 | DDX5 | DDX5 Entrez, Source | DEAD (Asp-Glu-Ala-Asp) box polypeptide 5 | 8037 | -0.031 | 0.1030 | No |
| 93 | CITED1 | CITED1 Entrez, Source | Cbp/p300-interacting transactivator, with Glu/Asp-rich carboxy-terminal domain, 1 | 8127 | -0.033 | 0.0983 | No |
| 94 | ARHGEF2 | ARHGEF2 Entrez, Source | rho/rac guanine nucleotide exchange factor (GEF) 2 | 8130 | -0.033 | 0.1001 | No |
| 95 | GRM2 | GRM2 Entrez, Source | glutamate receptor, metabotropic 2 | 8137 | -0.033 | 0.1016 | No |
| 96 | ESRRG | ESRRG Entrez, Source | estrogen-related receptor gamma | 8140 | -0.033 | 0.1035 | No |
| 97 | PITX2 | PITX2 Entrez, Source | paired-like homeodomain transcription factor 2 | 8349 | -0.037 | 0.0900 | No |
| 98 | CAPN6 | CAPN6 Entrez, Source | calpain 6 | 8491 | -0.039 | 0.0817 | No |
| 99 | HSD17B8 | HSD17B8 Entrez, Source | hydroxysteroid (17-beta) dehydrogenase 8 | 8628 | -0.041 | 0.0738 | No |
| 100 | SLIT3 | SLIT3 Entrez, Source | slit homolog 3 (Drosophila) | 8691 | -0.042 | 0.0717 | No |
| 101 | NR5A2 | NR5A2 Entrez, Source | nuclear receptor subfamily 5, group A, member 2 | 8704 | -0.042 | 0.0734 | No |
| 102 | LMO2 | LMO2 Entrez, Source | LIM domain only 2 (rhombotin-like 1) | 8708 | -0.042 | 0.0757 | No |
| 103 | RAB15 | RAB15 Entrez, Source | RAB15, member RAS onocogene family | 8760 | -0.043 | 0.0745 | No |
| 104 | PACSIN3 | PACSIN3 Entrez, Source | protein kinase C and casein kinase substrate in neurons 3 | 8824 | -0.044 | 0.0724 | No |
| 105 | PDYN | PDYN Entrez, Source | prodynorphin | 8860 | -0.045 | 0.0725 | No |
| 106 | MYST3 | MYST3 Entrez, Source | MYST histone acetyltransferase (monocytic leukemia) 3 | 9001 | -0.047 | 0.0647 | No |
| 107 | TRPC4 | TRPC4 Entrez, Source | transient receptor potential cation channel, subfamily C, member 4 | 9085 | -0.049 | 0.0614 | No |
| 108 | PLUNC | PLUNC Entrez, Source | palate, lung and nasal epithelium carcinoma associated | 9196 | -0.050 | 0.0561 | No |
| 109 | PPP1R10 | PPP1R10 Entrez, Source | protein phosphatase 1, regulatory subunit 10 | 9241 | -0.051 | 0.0559 | No |
| 110 | VIP | VIP Entrez, Source | vasoactive intestinal peptide | 9285 | -0.052 | 0.0559 | No |
| 111 | ESR2 | ESR2 Entrez, Source | estrogen receptor 2 (ER beta) | 9538 | -0.056 | 0.0402 | No |
| 112 | NKX2-5 | NKX2-5 Entrez, Source | NK2 transcription factor related, locus 5 (Drosophila) | 9922 | -0.065 | 0.0151 | No |
| 113 | MYH11 | MYH11 Entrez, Source | myosin, heavy chain 11, smooth muscle | 10036 | -0.067 | 0.0107 | No |
| 114 | MMP3 | MMP3 Entrez, Source | matrix metallopeptidase 3 (stromelysin 1, progelatinase) | 10103 | -0.068 | 0.0098 | No |
| 115 | MYH4 | MYH4 Entrez, Source | myosin, heavy chain 4, skeletal muscle | 10105 | -0.068 | 0.0139 | No |
| 116 | TOB1 | TOB1 Entrez, Source | transducer of ERBB2, 1 | 10294 | -0.072 | 0.0041 | No |
| 117 | PRPF4 | PRPF4 Entrez, Source | PRP4 pre-mRNA processing factor 4 homolog (yeast) | 10359 | -0.074 | 0.0037 | No |
| 118 | HAS2 | HAS2 Entrez, Source | hyaluronan synthase 2 | 10766 | -0.084 | -0.0219 | No |
| 119 | GJA5 | GJA5 Entrez, Source | gap junction protein, alpha 5, 40kDa (connexin 40) | 10784 | -0.085 | -0.0180 | No |
| 120 | FIP1L1 | FIP1L1 Entrez, Source | FIP1 like 1 (S. cerevisiae) | 10791 | -0.085 | -0.0133 | No |
| 121 | FGF23 | FGF23 Entrez, Source | fibroblast growth factor 23 | 10878 | -0.088 | -0.0145 | No |
| 122 | HMGN2 | HMGN2 Entrez, Source | high-mobility group nucleosomal binding domain 2 | 11136 | -0.094 | -0.0282 | No |
| 123 | GTF3C1 | GTF3C1 Entrez, Source | general transcription factor IIIC, polypeptide 1, alpha 220kDa | 11181 | -0.095 | -0.0258 | No |
| 124 | ATP5G2 | ATP5G2 Entrez, Source | ATP synthase, H+ transporting, mitochondrial F0 complex, subunit C2 (subunit 9) | 11226 | -0.096 | -0.0232 | No |
| 125 | MRPS18B | MRPS18B Entrez, Source | mitochondrial ribosomal protein S18B | 11420 | -0.103 | -0.0316 | No |
| 126 | AQP8 | AQP8 Entrez, Source | aquaporin 8 | 11422 | -0.103 | -0.0254 | No |
| 127 | SLC12A2 | SLC12A2 Entrez, Source | solute carrier family 12 (sodium/potassium/chloride transporters), member 2 | 11551 | -0.108 | -0.0286 | No |
| 128 | ABCA1 | ABCA1 Entrez, Source | ATP-binding cassette, sub-family A (ABC1), member 1 | 11627 | -0.110 | -0.0275 | No |
| 129 | RTN1 | RTN1 Entrez, Source | reticulon 1 | 11850 | -0.119 | -0.0371 | No |
| 130 | AMY2A | AMY2A Entrez, Source | amylase, alpha 2A; pancreatic | 11899 | -0.122 | -0.0333 | No |
| 131 | WDR20 | WDR20 Entrez, Source | WD repeat domain 20 | 11984 | -0.126 | -0.0320 | No |
| 132 | MYL2 | MYL2 Entrez, Source | myosin, light chain 2, regulatory, cardiac, slow | 12047 | -0.129 | -0.0289 | No |
| 133 | NEUROD1 | NEUROD1 Entrez, Source | neurogenic differentiation 1 | 12187 | -0.136 | -0.0312 | No |
| 134 | BIN1 | BIN1 Entrez, Source | bridging integrator 1 | 12208 | -0.137 | -0.0243 | No |
| 135 | GCH1 | GCH1 Entrez, Source | GTP cyclohydrolase 1 (dopa-responsive dystonia) | 12523 | -0.156 | -0.0386 | No |
| 136 | TGFBR1 | TGFBR1 Entrez, Source | transforming growth factor, beta receptor I (activin A receptor type II-like kinase, 53kDa) | 12538 | -0.158 | -0.0301 | No |
| 137 | NDNL2 | NDNL2 Entrez, Source | necdin-like 2 | 12742 | -0.177 | -0.0347 | No |
| 138 | EGF | EGF Entrez, Source | epidermal growth factor (beta-urogastrone) | 12819 | -0.188 | -0.0290 | No |
| 139 | CA2 | CA2 Entrez, Source | carbonic anhydrase II | 12972 | -0.213 | -0.0275 | No |
| 140 | SSBP3 | SSBP3 Entrez, Source | single stranded DNA binding protein 3 | 12979 | -0.214 | -0.0150 | No |
| 141 | PA2G4 | PA2G4 Entrez, Source | proliferation-associated 2G4, 38kDa | 13074 | -0.242 | -0.0074 | No |
| 142 | GADD45G | GADD45G Entrez, Source | growth arrest and DNA-damage-inducible, gamma | 13302 | -0.454 | 0.0030 | No |