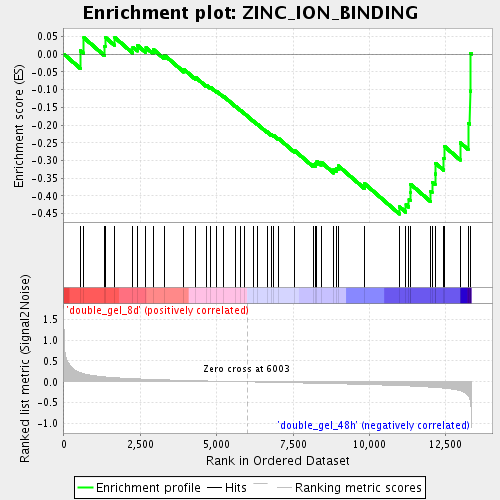
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | double_gel_and_single_gel_rma_expression_values_collapsed_to_symbols.class.cls #double_gel_8d_versus_double_gel_48h.class.cls #double_gel_8d_versus_double_gel_48h_repos |
| Phenotype | class.cls#double_gel_8d_versus_double_gel_48h_repos |
| Upregulated in class | double_gel_48h |
| GeneSet | ZINC_ION_BINDING |
| Enrichment Score (ES) | -0.45116293 |
| Normalized Enrichment Score (NES) | -1.5747449 |
| Nominal p-value | 0.011415525 |
| FDR q-value | 0.158301 |
| FWER p-Value | 1.0 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | GDA | GDA Entrez, Source | guanine deaminase | 537 | 0.219 | 0.0099 | No |
| 2 | RASSF1 | RASSF1 Entrez, Source | Ras association (RalGDS/AF-6) domain family 1 | 652 | 0.196 | 0.0465 | No |
| 3 | TIMM8B | TIMM8B Entrez, Source | translocase of inner mitochondrial membrane 8 homolog B (yeast) | 1334 | 0.117 | 0.0222 | No |
| 4 | PHEX | PHEX Entrez, Source | phosphate regulating endopeptidase homolog, X-linked (hypophosphatemia, vitamin D resistant rickets) | 1351 | 0.116 | 0.0477 | No |
| 5 | SIRT2 | SIRT2 Entrez, Source | sirtuin (silent mating type information regulation 2 homolog) 2 (S. cerevisiae) | 1665 | 0.099 | 0.0470 | No |
| 6 | LTA4H | LTA4H Entrez, Source | leukotriene A4 hydrolase | 2256 | 0.075 | 0.0200 | No |
| 7 | MSRB2 | MSRB2 Entrez, Source | methionine sulfoxide reductase B2 | 2405 | 0.071 | 0.0251 | No |
| 8 | CBLB | CBLB Entrez, Source | Cas-Br-M (murine) ecotropic retroviral transforming sequence b | 2684 | 0.063 | 0.0186 | No |
| 9 | CD4 | CD4 Entrez, Source | CD4 molecule | 2921 | 0.057 | 0.0139 | No |
| 10 | MMP12 | MMP12 Entrez, Source | matrix metallopeptidase 12 (macrophage elastase) | 3285 | 0.048 | -0.0024 | No |
| 11 | CBLC | CBLC Entrez, Source | Cas-Br-M (murine) ecotropic retroviral transforming sequence c | 3927 | 0.034 | -0.0428 | No |
| 12 | SIRT6 | SIRT6 Entrez, Source | sirtuin (silent mating type information regulation 2 homolog) 6 (S. cerevisiae) | 4310 | 0.027 | -0.0653 | No |
| 13 | PEX12 | PEX12 Entrez, Source | peroxisomal biogenesis factor 12 | 4664 | 0.021 | -0.0871 | No |
| 14 | MMP14 | MMP14 Entrez, Source | matrix metallopeptidase 14 (membrane-inserted) | 4808 | 0.018 | -0.0936 | No |
| 15 | MEP1B | MEP1B Entrez, Source | meprin A, beta | 4999 | 0.015 | -0.1045 | No |
| 16 | DUSP12 | DUSP12 Entrez, Source | dual specificity phosphatase 12 | 5220 | 0.011 | -0.1184 | No |
| 17 | MNAT1 | MNAT1 Entrez, Source | menage a trois homolog 1, cyclin H assembly factor (Xenopus laevis) | 5616 | 0.005 | -0.1468 | No |
| 18 | MMP2 | MMP2 Entrez, Source | matrix metallopeptidase 2 (gelatinase A, 72kDa gelatinase, 72kDa type IV collagenase) | 5780 | 0.003 | -0.1584 | No |
| 19 | MMP8 | MMP8 Entrez, Source | matrix metallopeptidase 8 (neutrophil collagenase) | 5909 | 0.001 | -0.1677 | No |
| 20 | MT3 | MT3 Entrez, Source | metallothionein 3 (growth inhibitory factor (neurotrophic)) | 6193 | -0.003 | -0.1883 | No |
| 21 | RNF4 | RNF4 Entrez, Source | ring finger protein 4 | 6343 | -0.005 | -0.1983 | No |
| 22 | MMP10 | MMP10 Entrez, Source | matrix metallopeptidase 10 (stromelysin 2) | 6646 | -0.010 | -0.2188 | No |
| 23 | BRCA1 | BRCA1 Entrez, Source | breast cancer 1, early onset | 6790 | -0.012 | -0.2268 | No |
| 24 | CTCF | CTCF Entrez, Source | CCCTC-binding factor (zinc finger protein) | 6862 | -0.013 | -0.2291 | No |
| 25 | BIRC5 | BIRC5 Entrez, Source | baculoviral IAP repeat-containing 5 (survivin) | 7015 | -0.015 | -0.2371 | No |
| 26 | SOD1 | SOD1 Entrez, Source | superoxide dismutase 1, soluble (amyotrophic lateral sclerosis 1 (adult)) | 7545 | -0.023 | -0.2715 | No |
| 27 | CSRP1 | CSRP1 Entrez, Source | cysteine and glycine-rich protein 1 | 8162 | -0.033 | -0.3102 | No |
| 28 | RPS29 | RPS29 Entrez, Source | ribosomal protein S29 | 8242 | -0.035 | -0.3082 | No |
| 29 | HDAC10 | HDAC10 Entrez, Source | histone deacetylase 10 | 8278 | -0.035 | -0.3027 | No |
| 30 | TIMM9 | TIMM9 Entrez, Source | translocase of inner mitochondrial membrane 9 homolog (yeast) | 8417 | -0.038 | -0.3045 | No |
| 31 | SLU7 | SLU7 Entrez, Source | - | 8831 | -0.044 | -0.3253 | No |
| 32 | S100B | S100B Entrez, Source | S100 calcium binding protein B | 8923 | -0.046 | -0.3216 | No |
| 33 | LNPEP | LNPEP Entrez, Source | leucyl/cystinyl aminopeptidase | 8978 | -0.047 | -0.3149 | No |
| 34 | ECE2 | ECE2 Entrez, Source | endothelin converting enzyme 2 | 9842 | -0.063 | -0.3653 | No |
| 35 | TIMM13 | TIMM13 Entrez, Source | translocase of inner mitochondrial membrane 13 homolog (yeast) | 10984 | -0.090 | -0.4305 | Yes |
| 36 | PIAS2 | PIAS2 Entrez, Source | protein inhibitor of activated STAT, 2 | 11194 | -0.096 | -0.4242 | Yes |
| 37 | TIMM10 | TIMM10 Entrez, Source | translocase of inner mitochondrial membrane 10 homolog (yeast) | 11292 | -0.098 | -0.4090 | Yes |
| 38 | TP53 | TP53 Entrez, Source | tumor protein p53 (Li-Fraumeni syndrome) | 11337 | -0.100 | -0.3893 | Yes |
| 39 | RPS27 | RPS27 Entrez, Source | ribosomal protein S27 (metallopanstimulin 1) | 11346 | -0.100 | -0.3669 | Yes |
| 40 | MMP16 | MMP16 Entrez, Source | matrix metallopeptidase 16 (membrane-inserted) | 11993 | -0.126 | -0.3865 | Yes |
| 41 | CHD4 | CHD4 Entrez, Source | chromodomain helicase DNA binding protein 4 | 12051 | -0.129 | -0.3611 | Yes |
| 42 | MMP9 | MMP9 Entrez, Source | matrix metallopeptidase 9 (gelatinase B, 92kDa gelatinase, 92kDa type IV collagenase) | 12150 | -0.134 | -0.3377 | Yes |
| 43 | YY1 | YY1 Entrez, Source | YY1 transcription factor | 12166 | -0.135 | -0.3078 | Yes |
| 44 | KPNB1 | KPNB1 Entrez, Source | karyopherin (importin) beta 1 | 12430 | -0.150 | -0.2931 | Yes |
| 45 | PRDM4 | PRDM4 Entrez, Source | PR domain containing 4 | 12458 | -0.152 | -0.2603 | Yes |
| 46 | CA2 | CA2 Entrez, Source | carbonic anhydrase II | 12972 | -0.213 | -0.2499 | Yes |
| 47 | MDM4 | MDM4 Entrez, Source | Mdm4, transformed 3T3 cell double minute 4, p53 binding protein (mouse) | 13232 | -0.326 | -0.1945 | Yes |
| 48 | APOBEC1 | APOBEC1 Entrez, Source | apolipoprotein B mRNA editing enzyme, catalytic polypeptide 1 | 13290 | -0.414 | -0.1037 | Yes |
| 49 | CRIP2 | CRIP2 Entrez, Source | cysteine-rich protein 2 | 13307 | -0.468 | 0.0026 | Yes |